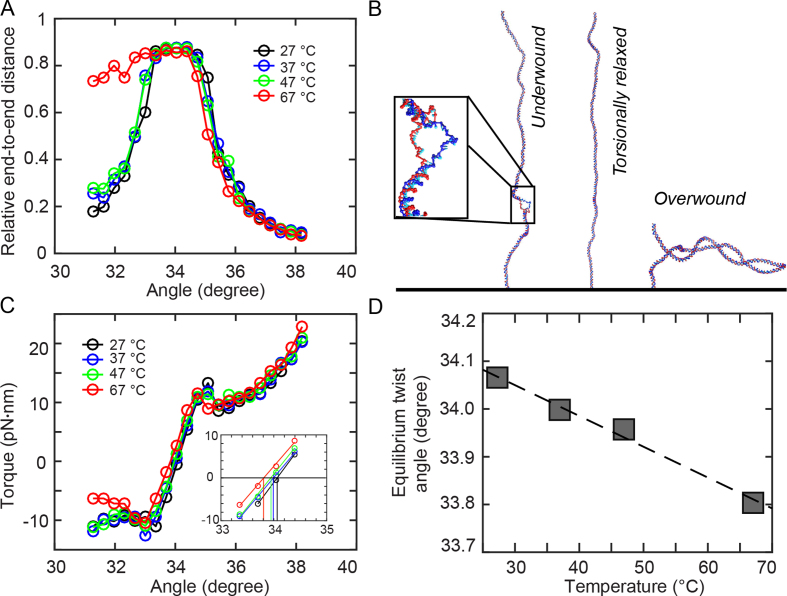
Figure 3.
Coarse-grained oxDNA simulations. Results from coarse-grained oxDNA simulations of a 600 bp double-stranded DNA system in the average-base parametrization at 150 mM monovalent salt. A stretching force F = 0.71 pN was used in all simulations. (A) Extension vs. imposed mean twist angle for 27, 37, 47 and 67°C. Note that for 67°C, denaturation bubble formation occurs in the undertwisted system (angles ≤ 33°), leading to asymmetric curves. (B) Molecular conformations observed in oxDNA simulations at T = 67°C for an angle of 31.6°, showing a denaturation bubble (left), 34.0°, showing an extended double-strand (center) and 37.2°, showing a plectoneme (right). For the 31.6° snapshot, a closeup of the 23-bp denaturation bubble is shown in the inset. (C) Torque response at different temperatures, showing a linear regime close to the equilibrium angle, followed by an overshoot and a post-buckling torque which increases more slowly upon further twisting. The inset shows a zoom of the data with the interpolation in the linear torque-regime and determination of the equilibrium angle corresponding to zero torque. (D) Equilibrium twist angles for the different temperatures determined by linear interpolation of the torque response shown in C to zero torque. The black line indicates a linear fit of temperature dependence, yielding ΔTw(T) = −6.5°/(°C·kbp).