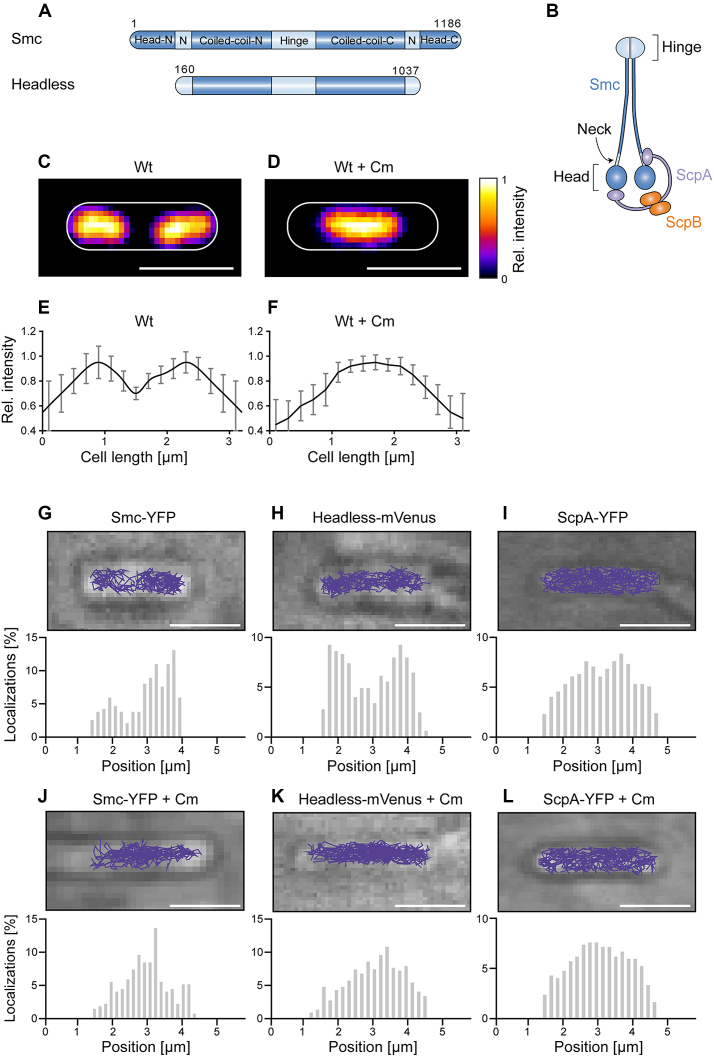
Figure 1.
(A) Domain organization of Smc and of headless Smc. Residue numbering corresponds to amino acids of Smc of Bacillus subtilis. Head-N: N-terminal part of head domain, N: Neck region, Coiled-coil-N: N-terminal coiled coil, Coiled-coil-C: C-terminal coiled coil, Head-C: C-terminal part of head domain. All proteins are tagged with the fluorescent proteins YFP or mVenus at the C-terminus. (B) The scheme indicates the organization of the complex of Smc, ScpA, and ScpB. (C) Typical image of a DAPI-stained cell of length ∼3.2 μm, (D) same as (C) after addition of chloramphenicol, (E and F) Distribution of normalized DAPI fluorescence in cells of ∼3.2 μm length in exponentially growing cells (n = 22) and in cells treated with chloramphenicol (n = 19) which results in a single compacted nucleoid in the cell middle. (G–L) The spatial organization of Smc and headless tracks reflects the organization of the nucleoid in exponentially growing B. subtilis cells, whereas ScpA tracks cover the entire cell. Upper panels show typical tracks obtained in a single cell, lower panels are histograms of localizations projected onto the long cell axis. Chloramphenicol treatment was used as a test for tracking on the nucleoid. Tracks were analyzed in cells of ∼3.2 μm length for better comparison.