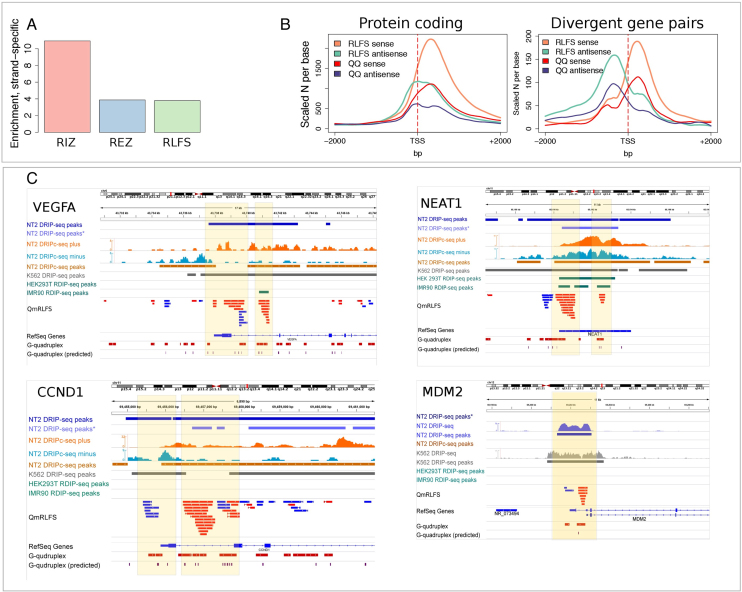
Figure 3.
RLFSs are co-enriched with experimental G4 sequences genome-wide. (A) Enrichment of experimental G4 structures on the sense RLFS strand. Enrichment was calculated as a ratio of number of the G4-positive merged RLFS sequences (RIZ, REZ or entire RLFS with at least one G4 on the sense strand) to the number of the G4-positive merged RLFS sequences (RIZ, REZ or entire RLFS with at least one G4, respectively) found on the same genome double strand position on the antisense strand. The strand orientation was defined by RLFS strand. (B) Distributions of RLFSs and experimental G4s in the proximal promoters (around TSS) of stand-alone protein-coding genes and the protein-coding/protein-coding divergent gene pairs. RLFSs were merged in a strand-specific manner to provide the same scaling with non-overlapping G4. (C) Genome browser shots showing RNA:DNA hybrids/R-loops and G4s in VEGFA, NEAT1, CCND1 and MDM2 gene promoters. Asterisk for DRIP-seq data denotes data from (46). Detailed descriptions of the maps and associations are presented in Supplementary Materials: examples of the RLFSs highly enriched with G repeats strand-specific G4-quadruplexes.