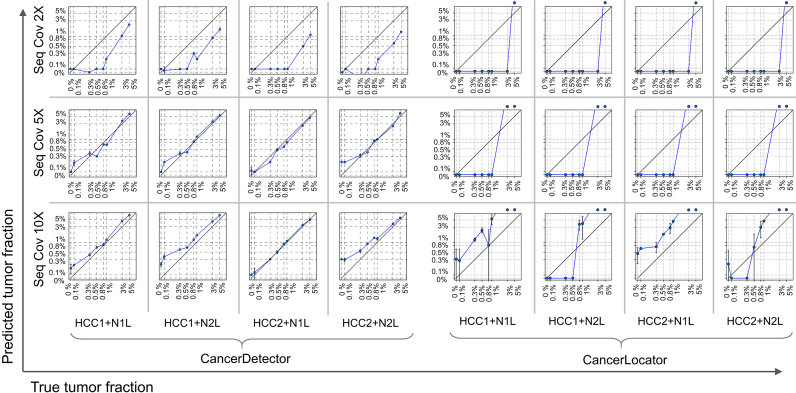
Figure 4.
Predicted blood tumor fractions (averaged over 10 runs) for the liver cancer cfDNA samples, simulated by subsampling and mixing sequencing reads from a real healthy cfDNA sample (N1L or N2L) and a solid liver tumor sample (HCC1 or HCC2) at eight different tumor fractions: 0, 0.1%, 0.3%, 0.5%, 0.8%, 1%, 3%, 5%, and at 3 different sequencing coverages (2×, 5× and 10×). In each log-log plot, a blue point represents a simulated sample with error bars (standard deviation of predicted tumor fraction), the x-axis is its true tumor fraction and the y-axis is its predicted tumor fraction. When the predicted tumor fraction is out of range (>5%), we draw the point above the box.