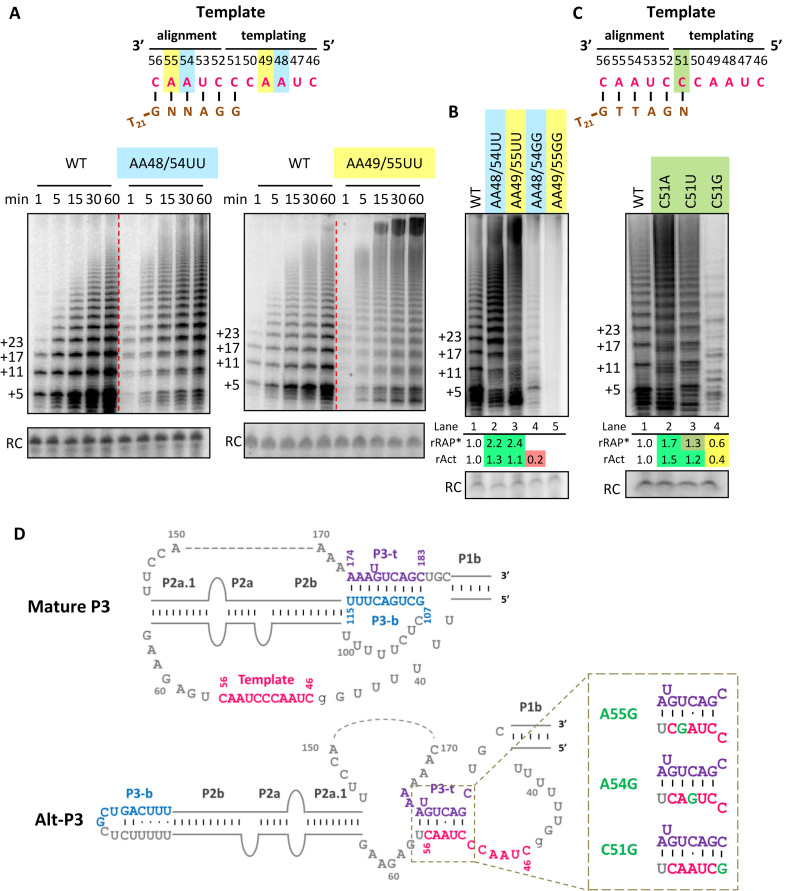
Figure 3.
Template mutations at the same position have different impact on activity and RAP of hTRmin telomerase assembled in vitro. (A) Activity assay of hTRmin mutants with A to U template mutations reconstituted in RRL, assayed over a time course in reactions omitting dCTP. Template sequence is shown at top, with shading indicating matching alignment and templating positions that pair to the same position of a telomeric repeat. Primer is shown in brown; N indicates a base complementary to the altered template. (B) Comparison of enzyme activity for templates with a pair of A mutated to U versus G, assayed in the presence of all dNTPs. (C) Activity assay of enzymes with mutations at the C51 template position, with an illustration at top parallel to (A). rRAP* values were calculated by dividing the collective intensities of products ≥10 repeats by the collective intensities of products <10 repeats and then normalizing the ratio to WT hTRmin enzyme. rAct values were calculated as described in Figure 1. For (B) and (C), similar results were obtained from two to three independent experimental replicates. (D) Two structures of the hTR t/PK with coloring to indicate the template (pink) and P3 strands top (P3-t, purple) and bottom (P3-b, blue). Active ‘Mature P3’ conformation has a template accessible for pairing with substrate, whereas Alt-P3 does not (adapted from (36)). At right, altered template sequences predicted to additionally stabilize alt-P3 are shown with the template mutation in green.