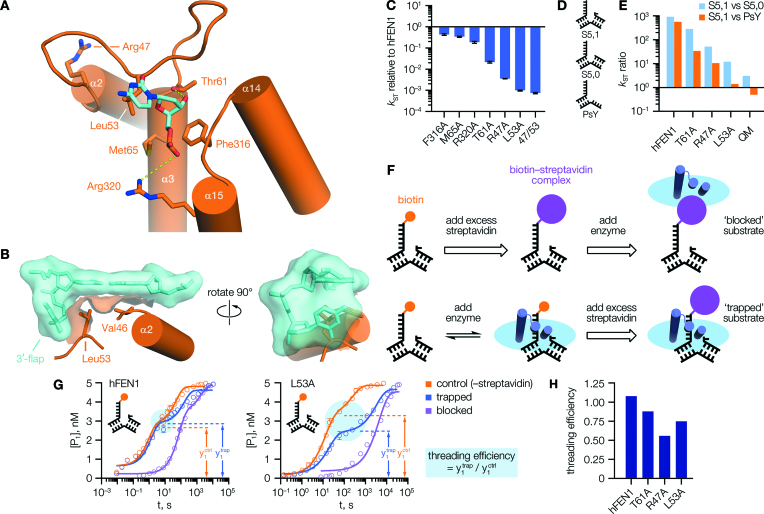
Figure 2.
3′-Flap binding mutations impaired FEN1 activity and threading stability. (A) Interactions made with the 3′-flap in an hFEN1–DNA complex (PDB code 5UM9 (10)). (B) Partial surface render of the same structure, showing the sidechain of Leu53 positioned as a ‘wedge’ contacting both the 3′-flap base, and the terminal base pair of the ‘upstream’ duplex. (C) Mutation of residues shown in (A) affected FEN1 single-turnover activity to varying degrees. (D) Structures of DNA constructs. (E) Mutations affecting 3′-flap binding impaired the enzyme's selectivity. (F) Schematic of threading assay, illustrating how formation of a streptavidin–biotin complex at the 5′-flap terminus was used to ‘lock’ DNA substrate in a threaded or unthreaded state. To prevent reaction, complexes were formed in Ca2+ buffer instead of Mg2+. (G) With hFEN1 (left), the preformed ‘trapped’ complex decayed to the same initial endpoint (cyan circle) as free substrate on addition of Mg2+, implying the labelled substrate was fully threaded at equilibrium. With hFEN1-L53A (right), these reactions reached different endpoints indicating some substrate was unbound/unthreaded at equilibrium. The apparent hFEN1 ‘control’ rate was slowed by the requirement to displace Ca2+ by Mg2+ here; see Supplementary Figure S7C and D for details. (H) Threading efficiency derived for various proteins. In (C), N = 2, n = 4 (F316A, M65A, R320A, L53A quench-flow phase); N = 2, n = 6 (T61A, R47A); N = n = 4 (L53A manual sampling phase). In (G), N = 2, n = 4 (quench-flow phase); for manual sampling phase, N = n = 4 (L53A) or 6 (hFEN1). Ratios in (E, H) are derived from data detailed fully in Supplementary Tables S4 and S5. Error bars show SEM.