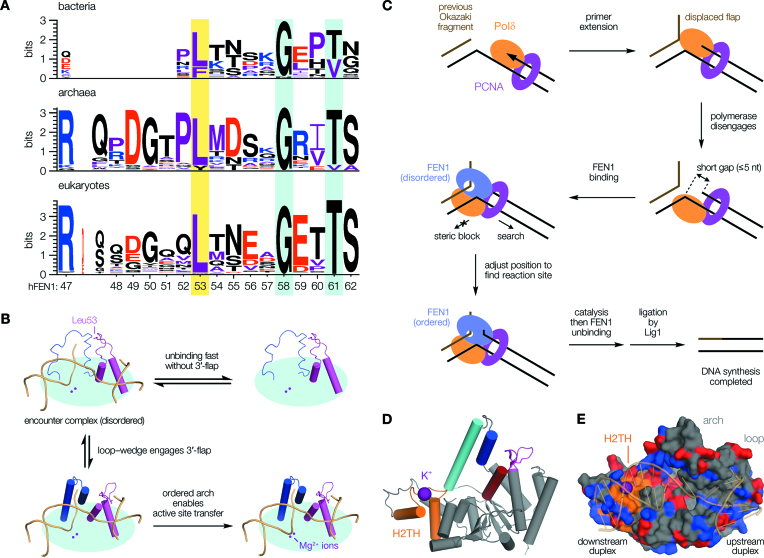
Figure 6.
Sequence analysis informs a refined model of cellular FEN1 function. (A) The wedge residue (yellow highlight) is present across all domains of life. Other loop positions discussed are also highlighted (cyan). (B) Engagement of a single nucleotide 3′-flap by the loop–wedge directly influences active site transfer, occurring over 20 Å away. (C) Model for FEN1 flap processing following strand displacement synthesis. Disengagement of DNA polymerase leaves a ‘short gap’ structure (42). Recruitment of FEN1 leads to rapid association with the downstream duplex via its H2TH domain, initiating a short-range ‘search’ to adjust the enzyme's position for reaction. Detection of the upstream (primer) duplex occurs through breaking the terminal base pair and binding the newly-formed 3′-flap, allosterically activating a cascade leading to catalysis. This concept applies equally whether or not any gap re-annealing occurs, requiring a minimum FEN1 translocation of one nucleotide (see Figure 4). It is likely inconsequential at what stage 5′-flap threading occurs during encounter, because this is fast relative to ordering for the short flap-lengths thought to predominate in vivo (56). Additionally, FEN1 and DNA ligase 1 (Lig1)—which seals the nick—cannot be co-resident on PCNA, with their switchover likely controlled through post-translational modifications (57–59). (D) hFEN1 structure, as Figure 1D, showing the H2TH domain (orange) and its bound K+ ion (purple). (E) Surface render of hFEN1 (PDB code: 3Q8K (26)) showing substrate DNA (transparent cartoon) tracking a channel of positively-charged sidechains (blue, with negatively-charged sidechains red). This emphasizes the largely electrostatic nature of the interaction, as has been recognized before (7,60).