Abstract
Mononegaviruses, such as Ebola virus, encode an L (large) protein that bears all the catalytic activities for replication/transcription and RNA capping. The C-terminal conserved region VI (CRVI) of L protein contains a K-D-K-E catalytic tetrad typical for 2’O methyltransferases (MTase). In mononegaviruses, cap-MTase activities have been involved in the 2’O methylation and N7 methylation of the RNA cap structure. These activities play a critical role in the viral life cycle as N7 methylation ensures efficient viral mRNA translation and 2’O methylation hampers the detection of viral RNA by the host innate immunity. The functional characterization of the MTase+CTD domain of Sudan ebolavirus (SUDV) revealed cap-independent methyltransferase activities targeting internal adenosine residues. Besides this, the MTase+CTD also methylates, the N7 position of the cap guanosine and the 2’O position of the n1 guanosine provided that the RNA is sufficiently long. Altogether, these results suggest that the filovirus MTases evolved towards a dual activity with distinct substrate specificities. Whereas it has been well established that cap-dependent methylations promote protein translation and help to mimic host RNA, the characterization of an original cap-independent methylation opens new research opportunities to elucidate the role of RNA internal methylations in the viral replication.
INTRODUCTION
Infections by Ebola virus can cause severe to fatal haemorrhagic fevers. The outbreak in West Africa in 2015 was unprecedentedly devastating causing more than 11,000 deaths (1). Despite huge efforts and investment in research for the development of countermeasures and for understanding the viral life cycle, Ebola is still a priority disease for the WHO, due to the risk of re-emergence and lack of efficient vaccines or antivirals. The genus Ebolavirus is comprised of five species: Zaire ebolavirus (Ebola virus, EBOV), Sudan ebolavirus (Sudan virus, SUDV), Tai Forest ebolavirus (Tai Forest virus, TAFV), Bundibugyo ebolavirus (Bundibugyo virus, BDBV) and Reston ebolavirus (Reston virus, RESTV) (2,3). They belong to the Filoviridae family together with other filamentous viruses such as Marburg marburgvirus (Marburg virus, MARV), which also causes acute haemorrhagic fevers. Having a non-segmented negative strand RNA genome (NNS), the Filoviridae are part of the Mononegavirales order, which also contains the well-characterized non-pathogenic vesicular stomatitis virus (VSV), as well as important human pathogens such as measles, Hendra and rabies viruses (4).
Filoviruses have a common genetic and structural organization. Their genome (∼19 kb) encodes seven proteins, namely the nucleoprotein NP, viral proteins VP35 and VP40, the glycoprotein GP, viral proteins VP30 and VP24 and the ‘large’ L protein (5,6). The replication/transcription cycle is driven by the L protein, which is embedded in cytoplasmic inclusion bodies (7,8).
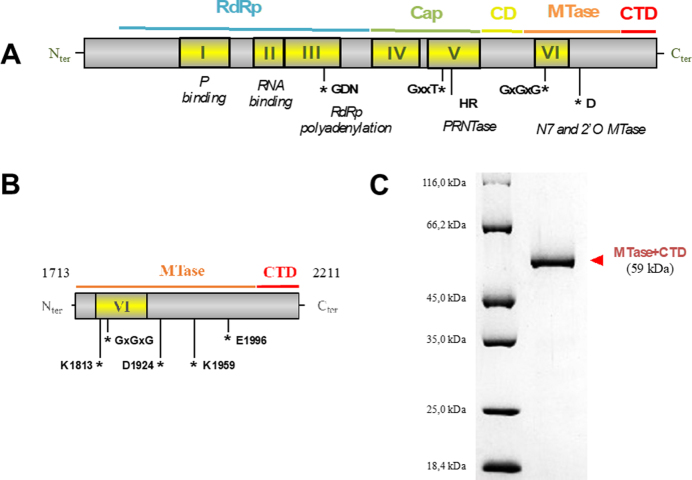
The L protein of mononegaviruses is a multi-functional protein. Although it varies in length, sequence analysis revealed that L protein contains six conserved regions (CRI to CRVI, Figure 1A) (9). To date, the only available structure of a complete L protein from a mononegavirus is that of VSV. The structure, determined by cryo-electron microscopy, revealed a topological organization in five domains (10) to which distinct functions were attributed. These are, from N- to the C-terminus (Figure 1A): the RNA-dependent RNA polymerase (RdRp) closely associated with a capping domain (Cap) containing polyribonucleotidyltransferase (PRNTase) activity, a structured connector domain (CD), the methyltransferase domain (MTase) and a small C-terminal domain (CTD). The enzymatic activities of the L protein are crucial for virus replication. Indeed, the RdRp domain drives genome replication and transcription into mRNAs (11), the PRNTase appends the cap structure onto the 5′ end of the viral transcripts (12–18) and the MTase is involved in methylation of the cap (19–23).
Figure 1.
The MTase+CTD domain of Sudan ebolavirus L protein. (A) Sequence analysis of the mononegavirus L protein revealed six conserved regions (CRI to CRVI, yellow boxes) that contain motifs responsible for the different activities of the L (motifs mapped with asterisks) (Poch et al., 1990). Additionally, the recently published VSV L structure resolved by cryo-EM shows that the L protein is organized in 4 domains: the RNA-dependent RNA polymerase (RdRp) intricated with the polyribonucleotidyltransferase (PRNTase or Cap), a connector domain, the methyltransferase domain (MTase) and a poorly conserved C-terminal domain (CTD). (B) Based on alignments with the VSV L protein, the MTase+CTD domain in SUDV L protein was defined as the fragment covering amino acids 1713–2211. SAM-binding site motifs (GxGxG) and the 2’O catalytic tetrad K-D-K-E have also been identified (asterisks). (C) SDS-PAGE of purified, recombinant SUDV MTase+CTD containing an N-terminal oligohistidine-tag (58.4 kDa).
The structure of the mRNA cap consists of a guanosine linked via a 5′-5′ triphosphate bridge to the 5′end of newly synthetized RNA transcripts, and the cap is methylated at the N7 position of this guanosine and at the 2’O position of the first (n1) residue (24). The cap is critical for virus replication as it protects viral mRNAs from cellular 5′ exonucleases, allows the recruitment of the cellular eIF4e factor for translation initiation, and hides viral RNAs from detection by innate immunity sensors such as RIG-I and MDA5 and interferon induced restriction factors such as IFIT molecules (for review, see (25)). The different enzymatic activities required for cap synthesis have already been characterized in mononegaviruses such as VSV (PRNTase and MTase activities) (17,22) and hMPV (MTase activity) (26). Strikingly, there is no biochemical or structural information on filovirus L proteins, likely because of difficulties in producing recombinant protein. Therefore, the capping process of filoviruses remained to be elucidated.
In this study, we produced and purified the C-terminal region of SUDV L protein and characterized its enzymatic activities. We showed that this domain has a dual MTase activity. We demonstrated that SUDV MTase predominantly carries out an unconventional, cap-independent 2’O-methylation targeting adenosine residues within capped and uncapped RNAs, in addition to the canonical cap-dependent N7 and 2’O methylations.
MATERIALS AND METHODS
Cloning and protein expression
Codon-optimized SUDV MTase+CTD synthetic genes (Biomers) were cloned into a pET14b vector for expression in bacteria. Mutations were introduced by PCR-amplifying the wild-type sequence using primers carrying the mutations, using the Turbo DNase (Ambion). PCR-products were purified using the Wizard SV PCR Clean-Up System (Promega). Transformed Escherichia coli T7 bacteria (NEB) were cultured at 30°C until an O.D. of 0.6 was reached, after which the temperature was changed to 17°C and IPTG (Sigma) was added to 20 μM. The next day, bacteria were spun down (8000 × g for 10 min at 4°C) using a Sorval Lynx 6000 centrifuge (Thermo), and pellets were stored at –80°C.
Purification of MTases
Pellets corresponding to 1 l of culture expressing SUDV MTase+CTD were thawed on ice, and bacteria lysed in a volume equal to 10 times final culture O.D. of optimized lysis buffer (1× BugBuster buffer (from a 10X solution, Merck) auditioned with 50 mM Tris pH 8, 150 mM NaCl, 5% glycerol, 30 mM imidazole, 1 mM PMSF, 100 μg/ml lysozyme, 1 μg/ml DNase and 1% Triton X100). After clarification (45 000 × g, 30 min, 4°C), lysates were incubated with CoNTA resin (Thermo; 0.5 ml/l culture) for 30 min at 4°C, with gentle shaking. The beads were transferred to a 25 ml column and washed with 2 × 20 ml of buffer W1 (50 mM Tris pH 8, 1 M NaCl, 5% glycerol, 30 mM imidazole) and 10 ml of buffer W2 (50 mM Tris pH 8, 150 mM NaCl, 5% glycerol, 150 mM arginine). Proteins were eluted in buffer E (50 mM Tris pH 8, 150 mM NaCl, 5% glycerol, 1 M arginine). Finally, proteins were concentrated using Amicon Ultra (EMD Millipore) ultrafiltration units, and stored at –80°C in 50% glycerol.
The coding sequence of the human RNA N7 MTase (RNMT), hMPV MTase, Zika virus (ZIKV) and dengue virus (DV) MTases were cloned in fusion with the coding sequence of a hexa-histidine tag, and corresponding recombinant proteins were expressed and purified as previously described (26–29). The vaccinia virus VP39 MTase was purchased (New England Biolabs).
Synthesis of RNA substrates
RNA sequences were chemically synthesized on a solid support using an ABI 394 synthesizer. After RNA elongation with 2′-O-pivaloyloxymethyl phosphoramidite monomers (30,31) (Chemgenes, USA), the 5′-hydroxyl group was phosphorylated and the resulting H-phosphonate derivative (32) was oxidized and activated into a phosphoroimidazolidate derivative to react with either pyrophosphate (giving pppRNA) or guanosine diphosphate (giving GpppRNA) (28,33). N7-methylation of the purified GpppRNA was performed enzymatically using N7-hMTase (28,33). To prepare monophosphate RNA (pG-SUDV12), the 5′-H-phosphonate RNA was treated with a mixture of N,O-bis-trimethylacetamide (0.4 ml), CH3CN (0.8 ml) and triethylamine (0.1 ml) at 35°C for 15 min, and then oxidized with a tert-butyl hydroperoxide solution (5–6 M in decane, 0.4 ml; 35°C, 15 min). After deprotection and release from the solid support with aqueous ammonia for 3 h at room temperature, RNA sequences were purified and validated to be >95% pure by IEX-HPLC and they were characterized by MALDI-TOF spectrometry.
To synthetize hairpin-structured RNAs, short RNAs from chemical synthesis were ligated to commercial complementary RNAs (Eurofins) with T4 RNA ligase 1 (New England Biolabs). Ligation reactions were realized in a total volume of 40 μl with 50 μM of both RNAs, 1 mM ATP and 0.5 U/μl of T4 RNA ligase 1 at 37°C for 2 h. RNAs were then purified with StrataClean beads (Agilent) to eliminate proteins and G25 column (GE Healthcare) to remove co-products and ATP excess. Ligation products were finally controlled on acrylamide gel and SafeView+ coloration (Abm).
In vitro transcription
Long synthetic RNAs have been produced by in vitro transcription using the HiScribe T7 High Yield RNA Synthesis Kit instructions (NEB), following the manufacturer's instructions. Templates are listed in Supplementary Table S2. Briefly, 1 μg of template was incubated with the T7 RNA polymerase mix and NTPs (40 mM) for 2 h at 37°C. Sodium acetate pH 5.2 and glycogen (Invitrogen) were added to the mix to 300 mM and 1 μg/μl, respectively, followed by 3 volumes of ethanol to precipitate the RNA. After centrifugation, pellets were washed with 70% ethanol and centrifuged again. Finally, pellets were dried and re-suspended in water.
MTase activity assay
To evaluate methyltransferase activities, a radioactive test was set up by mixing 4 μM of SUDV MTase+CTD domain with 1 μM of purified synthetic RNAs (see all RNAs in Supplementary Table S1), 10 μM of SAM and 0.5 μM of 3H-SAM (Perkin Elmer) in an optimized MTase assay buffer (50 mM Tris–HCl pH 8, 10 mM arginine). Reactions were stopped by a 10-fold dilution in water after 3 h at 30°C. Samples were transferred to DEAE filtermats (Perkin Elmer) using a Filtermat Harvester (Packard Instruments). Methyl transfer was then evaluated as described before (26).
Briefly, the RNA-retaining mats were washed twice with 10 mM ammonium formate pH 8, twice with water and once with ethanol. They were then soaked with liquid scintillation fluid (Perkin Elmer), allowing the measurement of 3H-methyl transfer to the RNA substrates using a Wallac MicroBeta TriLux Liquid Scintillation Counter13 (Perkin Elmer). The methyltransfer assay using the other viral MTases were performed in similar experimental conditions (50 mM Tris–HCl pH 8, 10 μM of SAM, 1 μM of purified synthetic RNAs). The enzyme concentrations used were the following: human RNA N7 MTase (RNMT) 0.1 μM, hMPV MTase 4 μM, ZIKV MTase 0.5 μM, DV MTase 0.5 μM and vaccinia virus MTase VP39 (2000 units, New England Biolabs).
Fluorescence polarization (FP)
Using T4 RNA ligase 1 (20 units; New England Biolabs), cyanine 5-cytidine-5′-phosphate-3′-(6-aminohexyl)phosphate (12.5 μM, Jena Bioscience) was ligated to the 3′ ends of the RNA substrates (10 μM) in T4 RNA ligase 1 buffer (New England Biolabs), 1 mM ATP (16°C, overnight). Ligase was removed by RNA precipitation in 3 M sodium acetate supplemented with glycogen (Thermo Scientific) (to 1 μg/μl). The fluorescent RNA was incubated (5 min at room temperature) with increasing concentrations of the SUDV MTase+CTD domain, in 50 mM Tris pH 8, 150 mM NaCl, 5% glycerol. Fluorescence polarization (FP) measurements were performed in a microplate reader (PHERAstar FS; BMG Labtech) with an optical module equipped with polarizers and using excitation and emission wavelengths of 590 and 675 nm, respectively. Dissociation constants (Kd) were determined using Hill slope curve fitting (Prism).
RNA digestion assay and HPLC analysis
Each HO-(A)27, HO-(Am)27 and HO-(A)27 RNA which was preliminarily treated with the SUDV Mtase+CTD domain (2.5 μg/μl) in buffer (50 mM Tris–HCl pH 7.5 containing 50 mM NaCl, 10 mM MgCl2; final concentrations) was incubated with snake venom phosphodiesterase (0.2 unit/μmol RNA) at 37°C for 15 h. Then alkaline phosphatase (150 units/μmol RNA) was added and the mixture was incubated at 37°C for a further 30 min. The resulting digestion mixture was then analyzed by reverse-phase HPLC (column Macherey-Nagel Nucleodur C18, 7.5 × 4.6 mm, 100-3 EC, flow rate: 1 ml/min). Elution was performed with a 20 min linear gradient of 8% of acetonitrile in 50 mM triethylammonium acetate, pH 7. UV detection was carried out at 260 nm.
Thin layer chromatography (TLC)
Radioactive capped RNAs (G*pppRNA) were synthesized by incubating pppRNA (10 μM) with vaccinia virus capping enzyme (New England Biolabs) in the presence of 1.65 mCi of [α-32P]-GTP (Perkin Elmer). Labeled RNAs were purified with StrataClean beads (Agilent) to remove proteins and G25 columns (GE Healthcare) to remove excess of radioactive GTP. RNA was then submitted to methylation by the SUDV MTase+CTD domain (as above), and precipitated as described for the in vitro transcription products. Finally, RNAs were digested with 1 U of nuclease P1 (Sigma) in 30 mM sodium acetate (pH 5.3), 5 mM ZnCl2 and 50 mM NaCl (4 h, 37°C). Products were spotted onto polyethylenimine cellulose thin-layer chromatography plates (Macherey-Nagel) and resolved using 0.65 M LiCl as mobile phase. The radiolabeled caps released by nuclease P1 were visualized using a Fluorescent Image Analyzer FLA3000 (Fuji) phosphor-imager.
RESULTS
The SUDV MTase+CTD domain exhibits an unconventional RNA methyltransferase activity
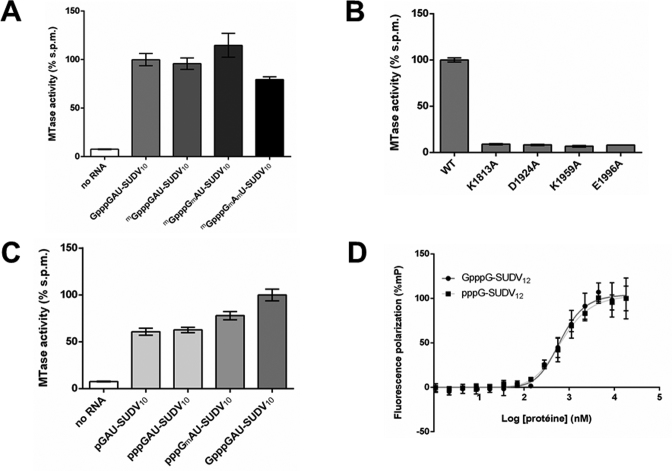
Based on bioinformatics analyses (Supplementary Figure S1), we designed several constructs encoding the C-terminal domains of SUDV L protein, and we tested their expression in bacteria. We selected the construct spanning amino-acids 1713–2211 (Figure 1B), which produced soluble MTase+CTD protein. After optimization of the solubilisation buffer, the SUDV MTase+CTD was purified to homogeneity as previously described for hMPV (26) (Figure 1C). To characterize its MTase activity we first monitored the transfer by the SUDV MTase+CTD domain of radioactive methyl groups from the S-adenosylmethionine (SAM) methyldonor to a variety of capped RNAs mimicking the conserved 5′ end of SUDV transcripts. Some of these RNAs contained non-radioactive methyl groups at key positions (i.e. the N7-position of the cap or the 2’O-positions of n1 or n2—see Supplementary Table S1), to help identify the positions to which the radiolabelled methyl groups were transferred (Figure 2A). Unexpectedly, we observed a strong MTase activity on cap-0 (mGpppG), cap-1 (mGpppGm) and cap-2 (mGpppGmAm) RNAs. This activity was not observed with human RNA-N7 methyltransferase (RNMT) or the vaccinia virus 2’O MTase (VP39), whose activities result in mG and n1m caps, respectively (Supplementary Figure S2A). The methylation profile is also different from that observed with the MTase+CTD domain of the closely related hMPV virus, which mainly targets the mRNA cap structure (Supplementary Figure S2B) and is weakly active on cap-1 RNA substrates. These observations thus suggest that the SUDV MTase+CTD methylates RNA at unconventional positions. To confirm the specificity of methylation observed, we mutated the conserved K-D-K-E catalytic residues typical for 2’O methyltransferases. Supplementary Figure S1 and Figure 2B demonstrated that MTase activity is strongly decreased by alanine mutation of each 2’O MTase catalytic residues. We then examined whether the presence of the cap was needed for this unconventional activity, and observed that both monophosphate and triphosphate RNAs could be methylated, although to a lower extent than the capped form, indicating cap-independent methylation in our experimental conditions (Figure 2C). This methylation does not seem to target the n1 as pppGmAU-SUDV9 is also methylated. These observations are further supported by similar dissociation constants measured for binding to pppG-SUDV12 and GpppG-SUDV12 (641 and 682 nM, respectively), suggesting SUDV MTase+CTD targets sites outside the cap (Figure 2D). Since the MTase activity depends on the 2’O MTase catalytic tetrad (Figure 2B), we examined whether the methylation reaction targets 2’OH groups of nucleotides within the RNA.
Figure 2.
SUDV MTase+CTD has a cap-independent MTase activity. The Sudan ebolavirus (SUDV) methyltransferase domain MTase+CTD domain exhibits methyltransferase activity. (A) SUDV MTase activity measurements on a set of synthetic RNAs (13-mers) corresponding to the 5′end of SUDV mRNAs, some of which are pre-methylated at specific positions (Gppp: cap, mGppp: N7-methylated cap, Gm: 2’O-methylated nucleotide). Results have been normalized to those obtained with an unmethylated control (n = 6). Values represent normalized mean ± standard deviation. (B) MTase activity measurements of SUDV catalytic wild-type (WT) and mutants with altered catalytic K-D-K-E tetrad residues on a capped synthetic SUDV sequence-specific RNA (Gppp-13 mers). Mutant activities were normalized to those of a WT control (n = 3). Values represent normalized mean ± standard deviation. (C) SUDV MTase activity measurements on a set of synthetic SUDV-sequence-based RNAs (13 mers) with differing 5′ ends (p: monophosphate, ppp: triphosphate, Gppp: cap). Results have been normalized to those obtained with an unmethylated control (n = 6). Values represent normalized mean ± standard deviation. (D) Fluorescence polarization measurements of the SUDV MTase+CTD domain for synthetic SUDV sequence-specific RNAs (13 mers) with or without a cap structure (Gppp: cap, ppp: uncapped) (n = 3). Kd values were very similar, 0.64 μM compared to 0.68 μM, respectively. Values were normalized to the highest signal for each RNA (n = 3) and are given as normalized mean ± standard deviation.
The unconventional activity of SUDV MTase+CTD specifically targets adenosines within RNA substrates
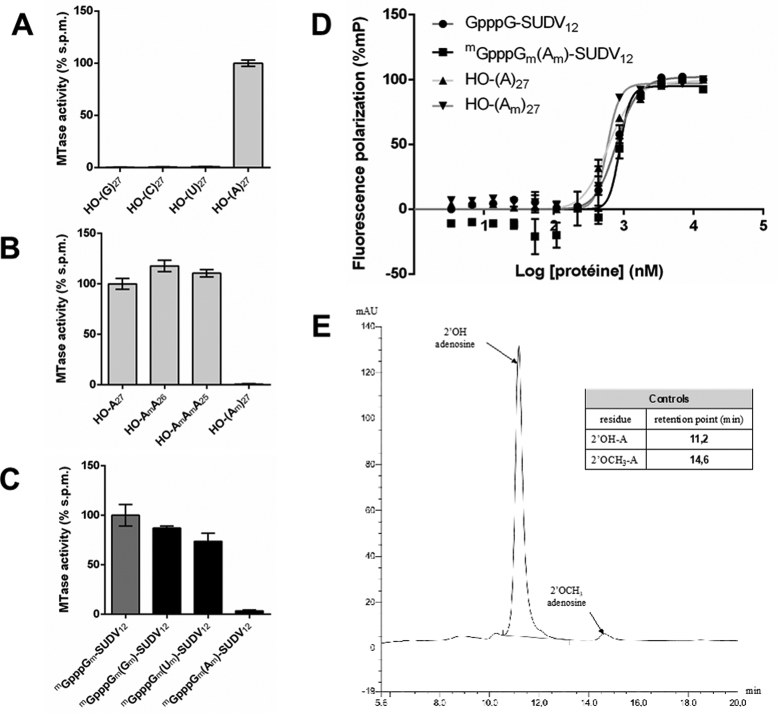
By using various homopolymeric RNA substrates (27-mers), we demonstrated that the MTase+CTD preferentially methylates adenosine residues (Figure 3A) and is not active on poly G, C and U RNAs. Conversely, the MTase+CTD was unable to transfer methyl groups to the A27 RNA (HO-(Am)27) substrate, of which all adenosines were pre-methylated at their 2’OH groups (Figure 3B). In addition, MTase activity is still detected using HO-(Am)A26 and HO-(AmAm)A25 RNAs as substrates. These results suggest that the SUDV MTase+CTD targets 2’O positions of adenosines within the RNA substrates downstream of the cap structure. This internal adenosine MTase activity appears to be the main activity of SUDV MTase in our experimental conditions. Conversely, the ratio of MTase activities on cap-1 RNAs (mGpppXm-RNA) over those on capped but unmethylated RNAs (GpppX-RNA) indicates that hMPV, ZIKV and DV MTases predominantly methylate the cap-structure (Supplementary Figure S3A). Additionally, SUDV MTase+CTD also methylates double-stranded RNAs, contrary to ZIKV MTase that only targets adenosines of single-stranded RNAs (Supplementary Figure S3B) (29). Further tests showed that heteropolymeric cap-1 RNAs mimicking the conserved start sequence of SUDV 5′ transcripts are good MTase substrates even if their inner G or U residues were previously methylated (Figure 3C). Conversely, prior 2’O methylation of adenosine residues almost completely abolishes methyl transfer, confirming that there is no other unconventional activity on the cap structure. Methylation of internal residues does not decrease RNA/protein affinity as Kds for GpppG-SUDV12, mGpppGm(Am)-SUDV11, HO-(A)27 and HO-(Am)27 are similar (796, 881, 610 and 582 nM, respectively). To confirm that SUDV MTase is indeed a 2’O MTase, we analysed by HPLC the resulting digestion mixture of HO-(A)27 RNA preliminarily methylated by SUDV MTase+CTD domain upon degradation with a 3′-exonuclease and alkaline phosphatase (Figure 3E). After digestion, 98% of released residues correspond to native unmethylated adenosines and the remaining 2% were assigned to 2’O-methyl adenosines (Supplementary Figure S4), which is consistent with the ratio of not transferred versus transferred tritiated methyl group during MTase assay (Supplementary Figure S5). These data confirm the identification of an internal adenosine-specific 2’O methyltransferase activity (internal A-2’O MTase).
Figure 3.
SUDV MTase+CTD methylates internal adenosine residues of synthetic RNAs. The Sudan ebolavirus (SUDV) L protein MTase with the C-terminal domain (MTase+CTD) catalyses internal methylations. (A) MTase activity measurements on synthetic, 27-nucleotide-long homopolymeric RNAs (HO-(G/C/U/A)27). Groups have been normalized versus the activity on HO-(A)27 (n = 6). Values represent normalized mean ± standard deviation. (B) MTase activity evaluation on synthetic HO-(A)27 RNAs with 2’O-methylated residues (Am) or all residues 2’O methylated ((Am)). Groups have been normalized versus the activity on HO-(A)27 (n = 6). Data represent normalized mean ± standard deviation. (C) MTase activity evaluation on a set of synthetic SUDV sequence-specific RNAs (13 mers) with cap-1 (mGpppGm) and internal 2’O-methylated residues (Gm/Um/Am)). Groups have been normalized versus the activity on SUDV sequence-specific RNA with a cap-1 and no internal methylation (n = 6). Data represent normalized mean ± standard deviation. (D) RNA affinity evaluation by fluorescence polarization of synthetic SUDV sequence-specific capped RNAs (13-mers) with or without a cap-1 (mGpppGm) and internal 2’O-methylated adenosine residues (Am) and polyadenosine RNAs with or without 2’O methylations for the SUDV MTase+CTD domain (n = 3). Respective Kd are estimated at 0.88, 0.80, 0.58 and 0.61 μM. Results have been normalized versus the highest signal for each RNA (n = 3). Data represent normalized mean ± standard deviation. (E) HPLC profile of SUDV MTase+CTD methylated HO-(A)27 following treatment with snake venom phosphodiesterase and alkaline phosphatase. Controls identify adenosine (2’OH-A) at a retention point of 11.2 min and 2’O-methylated adenosine (2’OCH3-A) at a retention point of 14.6 min. These compounds were detected in a 98:2 ratio in the methylated HO-(A)27 RNA.
The SUDV MTase+CTD domain also shows cap methyltransferase activity
To further characterize cap methylations, we used synthetic RNAs with internal 2’O-methylated adenosines and caps pre-methylated at different positions ((m)GpppX(m)(Am)-SUDV11). These RNAs mimicking the conserved start sequence of SUDV 5′ transcripts and start with either an A or a G. Only the cap with a 2’O-methylated G (GpppGm) at n1 was specifically methylated (Figure 4A), indicating that SUDV MTase+CTD has a cap N7 MTase activity, which is dependent on prior 2’O methylation of the n1 guanosine. This N7 MTase activity is pH-dependent with an optimal pH ranging between 7.0 and 7.5 whereas 2’O internal methylation peaks at a higher pH (Supplementary Figure S6). We further characterized the nature of the methylation of the cap structure by TLC analysis (Figure 4B). In this time course experiment, 2’O-methylated RNAs with 32P-radiolabeled caps were incubated with SUDV MTase+CTD before RNA digestion by P1 nuclease. The release of a cap-1 (mGpppGm) structure could be evidenced by TLC separation (Figure 4B). Altogether, these experiments demonstrate that the MTase+CTD domain of SUDV has cap N7-MTase activity on RNAs starting with a G. Interestingly, we did not detect N7 methylation using cap 2’O-methylated RNA beginning with an A (GpppAm), suggesting substrate specificity based on the first nucleotide of the RNA. While this methylation requires a 2’O-methylated nucleotide at the first position of the RNA, cap 2’O methylation could not be detected in our enzymatic assays performed with short RNA substrate (VP40-13 mers). We thus evaluated cap-dependent 2’O MTase activity on synthetic RNA substrates of various lengths mimicking the 5′end of VP40 mRNA (Supplementary Tables S1 and S2). The short RNAs (VP40-13-mers) could not be methylated (Figure 4C), confirming our previous results. However, longer RNAs (VP40-31-mers and longer (not shown)) became 2’O-methylated on the n1 guanosine (Figure 4C), suggesting that RNA length or folding is a key factor for cap-dependent 2’O methylations.
Figure 4.
SUDV methylates its cap structure on the N7 position. The Sudan ebolavirus (SUDV) L protein MTase with the C-terminal domain (MTase+CTD) catalyses internal methylations and also methylates the cap structure. (A) MTase activity measurement at pH 7.0 and 8.0 on a set of synthetic SUDV sequence-specific RNAs (13-mers) that are uncapped (pppG) or capped (GpppX, mGpppX, GpppXm, mGpppXm), and carry internal 2’O-methylated adenosine residues ((Am)). Results have been normalized versus the activity on GpppG(Am)-SUDV12 RNA (n = 3). Values represent normalized mean ± standard deviation. (B) Thin layer chromatography of cap structures of control RNAs and a synthetic SUDV sequence-specific RNA (13-mers) with a 2’O-methylated cap (GpppGm) and internal 2’O methylated adenosines (Am) incubated with SUDV MTase+CTD domain for 0h, 3h and overnight (O/N). (C) Thin layer chromatography of cap structures from control RNAs and from synthetic SUDV VP40 sequence-specific RNAs (13-, 31-mers) with an unmethylated cap (GpppG). All RNAs were incubated with the SUDV MTase+CTD domain for 6h.
DISCUSSION
To date, there are few structural or biochemical data on the Ebola virus L protein, which compromises our understanding of the viral replication/transcription mechanisms and development of inhibitors with potential antiviral effect. In this study, we produced and purified the C-terminal region of the SUDV L protein encompassing the MTase and CTD domains (MTase+CTD). The functional study of this protein leads to the discovery of three MTase activities allowing the N7 and 2’O methylations of RNA cap structure and 2’O methylation of internal adenosines within RNA. Internal A-2’O MTase activity has not yet been reported in Mononegavirales in contrast to flaviviruses (29,34) with a putative biological role on virus replication, transcription.
In agreement with previous studies on mononegavirus L proteins, the cap N7 methylation function was expected (22,26,35). Our results indicate that this cap methylation occurs only if the first RNA nucleotide is a 2’O-methyl guanosine, like hMPV and VSV capping systems where the 2’O methylation of the first nucleotide precedes the N7 methylation of the cap guanosine (22,26). Nevertheless, we did not detect any N7 methylation of capped RNAs starting with a 2’O-methyl adenosine in our experimental system, suggesting substrate specificity based on the first nucleotide of the RNA. Since EBOV mRNAs start with an n1 guanosine (Dr Roland K. Hartmann, personal communication), this sequence specificity would ensure the specific methylation of SUDV mRNA. Contrary to what was observed with other viruses, such as coronaviruses (36,37) or flaviviruses (38–40), the 2’O methylation of the n1 residue in SUDV is barely detected on short RNA and requires RNAs longer than 30 nucleotides. Interestingly, the addition of the cap in VSV requires RNAs that are at least 31 nucleotides long (18). These results prompt us to propose that transcription in the L protein starts off with the synthesis of a ∼30 nt-long uncapped RNA, which remains covalently bound to the PRNTase domain before the non-methylated cap guanosine is added. The RNA cap is then methylated at the 2’O position of n1 before N7 methyaltion of the cap guanosine by the MTase+CTD domain of L protein.
In addition to its cap-MTase activity, SUDV MTase harbors an unexpected MTase activity inducing viral epigenetic or epitranscriptomic RNA modifications on both capped and uncapped RNAs. We demonstrated that the SUDV MTase targets adenosines present within homo- and hetero-polymeric RNA sequences at the 2’O position of the ribose. Unlike the n1 guanosine, internal guanosines are not methylated suggesting that substrate recognition is different for cap-dependent and cap-independent 2’O methylations. Interestingly, the structure of the closely related hMPV MTase lacks a cap-binding site such as that present in flaviviruses, coronaviruses and vaccinia virus where the cap is stacked between aromatic residues (26,37,41,42). As SUDV MTase+CTD (like its hMPV counterpart) recognizes both cap and uncapped RNAs with a similar affinity and all SUDV MTase activities are carried out by a single catalytic site, our results suggest that the domain has evolved away from the requirement of a cap-binding site.
Different optimal pHs have been reported for N7 and 2’O MTase activities (41). The SUDV N7 MTase activity appears more efficient at pH 7–7.5, which is consistent with the optimal pH described for other viral N7 MTases such as the ones of VSV, SeV or flaviviruses (22,29,42). On the other hand, the SUDV 2’O MTase activity takes place within a broad pH range from 8 to 10 as previously described for VSV and flavivirus MTases (22,42). This difference suggests that N7 and 2’O-MTase activities have a different chemical mechanism to transfer the methyl group from the SAM molecule to the RNA. This hypothesis has already been proposed for flaviviruses (42,43), which are structurally similar to both the N7 MTase of the protozoon Encephalitozoon cuniculi (Ecm1) and the vaccinia virus VP39 2’O MTase (44,45). Yet, it has been shown that these two enzymes use two distinct mechanisms: N7 methylation in Ecm1 uses direct proximity and geometry of substrates (48) instead of a SN2 nucleophilic attack on the SAM following a 2’OH deprotonation for the 2’O methylation (46,47). Taken together, these results show that the overall MTase reaction mechanisms involving the K-D-K-E catalytic tetrad are conserved despite a putative absence of a cap-binding site, and that it is the manner at which RNA is presented to the active site, which determines whether cap or internal methylation takes place. The CTD domain of L protein may play a critical role in this process, as this basic domain is probably a key factor for recruitment and positioning the substrate RNA onto the active site of the MTase (47). It is thus possible that structural differences of CTDs among mononegaviruses may explain the different methylation activities observed.
Most viral MTases perform cap-dependent N7 and/or 2’O methylations (Supplementary Figure S2A) (34,48–51). However, it has recently been shown that flavivirus MTases (Dengue virus and Zika virus) additionally carry out internal A-2’O methylations (29,34). These activities might be related to the epitranscriptomic RNA modifications recently reported in the genome of flaviviruses (52). Here, we report for the first time that internal A-2’O MTase activity also exists in viruses belonging to the Mononegavirales order (i.e. hMPV and SUDV) (Figure 2 and Supplementary Figure S3A). The level of internal adenosine methylation obtained with the MTase+CTD of SUDV is clearly much higher than those obtained with hMPV, Zika and Dengue viruses, which predominantly target the cap structure. This higher level may be due to the fact that SUDV MTase is able to methylate A-2’O groups on both single- and double-stranded RNA structures, unlike ZIKV MTase, which carries out A-2’O methylations on single-stranded RNA only (Supplementary Figure S3B). It is interesting to note that SUDV and hMPV MTase induces mainly internal A-2’O methylation as recently described for ZIKV and dengue virus MTase. This observation suggests that residues in the RNA binding site specifically recognize adenosines residues. In the case of dengue virus MTase, the molecular basis of the specific adenosine recognition at n1 position was elucidated. The crystal structure of the dengue virus MTase in presence of its RNA substrate indicate that the n1 adenosine base fits in a pocket shaped by residues I147-G148-E149-S150 downstream of the D146 catalytic residue (53). The structure also reveals that G, in place of A residue would sterically clash with SAH and that pyrimidine (U or C) bases would decrease the van der Waals interactions with the protein explaining why the dengue virus MTase induces internal 2′-O-methylations on polyA but not on polyG, polyC, or polyU substrates (34). Unfortunately, the residues downstream of the D catalytic residues are not conserved in SUDV MTase, and it is impossible to infer the residue involved in A1 recognition in the absence of structural data.
Our work also indicates that the methyltransferase of Ebola virus has evolved to catalyze epigenetic or epitranscriptomic internal RNA 2’O-methylations in addition to the canonical cap methylations, and this to a much higher level than that of the other hMPV, illustrating a divergent evolution within the Mononegavirales order. Several studies recently reported the dynamics and complexity of viral epigenetic or epitranscriptomic RNA modifications during infection (52). Among them, internal A-2’O methylations are one of the most abundant (≈0.3%) although some of the viruses in the studies do not encode any MTase domain. It is noteworthy that, using SUDV MTase, we observed the same specificity of methylation on adenosine residues with ∼0.5% of A-2’O methylations. Additional work is now necessary to identify if host RNAs or viral RNAs are targeted by this viral MTase activity and to evaluate the importance of such RNA methylation in the context of innate immunity escape.
The function of such internal A-2’O methylations is not yet understood. Functional analysis using a flavivirus replicon showed that internal methylations attenuate viral RNA replication in BHK-21 cell lines (34). In addition, internal A-2’O methylations may reduce the efficiency of polymerisation in an in vitro RNA elongation assay (34). On the other hand, it is also possible that epigenetic/epitranscriptomic RNA modifications increase viral replication by limiting the detection of viral RNAs by intracellular sensors and/or restriction factors. Indeed, methylations contribute to structural diversity of RNAs and thus regulate recognition by host proteins (54). One other possibility is that internal A-2’O methylations would regulate encapsidation. In mononegaviruses, genomic and anti-genomic RNAs are encapsidated by a nucleoprotein (NP) but not mRNAs. Structural work (55) has shown that NP/RNA interactions involve 2’OH groups on the nucleotides. Internal methylation of these groups may thus help NP to discriminate mRNAs from genomic and anti-genomic RNAs.
RNA methylation also plays a key role in disguising viral RNAs as host ‘self’ RNAs, allowing them to escape host surveillance by intracellular sensors regulating interferon expression. Among them, Toll-like receptors and RIG-like cytosolic receptors such as RIG-I and MDA5 have been demonstrated to sense methylated RNA (56–59). Whereas RIG-I has been shown to detect 5′ends of mis-capped RNAs, MDA5 may play a key role in the detection of double-stranded RNA without internal methylations. It is also possible that interferon-stimulated genes (ISGs) or restriction factors could detect unmethylated RNAs as ‘non-self’, thereby limiting viral replication. Proteins from the IFIT family, for example, recognize the 5′end of triphosphate or mis-capped RNAs and block their translation into proteins (60,61). Another example is ADAR, a deaminase known to suppress filovirus replication by RNA editing (62,63), whose activity is hampered by the presence of 2′-OH methyl groups (64). A role of 2’O-methylation in shielding the virus from the host's innate immunity system could also explain why replication of an EBOV minigenome is not inhibited by type I, type II or type III interferons (65), even in the absence of VP24, which helps repress ISG expression (66,67). Future research is needed to further characterize the internal methylation of Ebola virus RNA and to elucidate its role in the live cycle and pathogenesis of the virus.
CONCLUSION
This study reports three distinct methyltransferase activities carried out by the C-terminal region of SUDV L protein. Two of them are the traditional, cap-dependent N7 and 2’O methyltransferase activities that complete the synthesis of the viral cap structure, mimicking that on host mRNAs, and enabling mRNA completion and translation. The third one, a cap-independent 2’O-A methyltransferase activity, was not described before, and may further promote virus survival, possibly by shielding the RNAs from intracellular sensors and restriction factors thus promoting immune evasion, or by regulating RNA encapsidation by the nucleoprotein. These methylations might thus be important during Ebola virus infection. Although it remains to be elucidated why this activity is specifically prevalent in Ebola viruses, it has clear potential as a potent antiviral target.
Supplementary Material
ACKNOWLEDGEMENTS
The research was made possible by funding from a DGA/Aix-Marseille Université PhD fellowship for B.M. B.M., E.D. and B.C. thank Dr Barbara Selisko, Dr Hervé Bourhy and Dr Jean-François Elëouet for their feedback and constructive remarks.
Author Contributions: B.M., B.Co. and E.D. conceived and designed the study. B.M. B.Co., T.G. and E.D. performed experiments, using material prepared by T.G., G.C.P. and F.D., B.M., B.Co., F.D. and E.D. analyzed data and wrote the paper. G.C.P., B.Ca., J.J.V. and J.M.G. revised the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Union Seventh Framework Programme [FP7/2007-2013] under SILVER grant agreement [260644]; MRC grant [MR/L017709/1]; WT [200835/Z/16/Z to J.M.G.]; Wellcome Trust administrative support grant [203141/Z/16/Z]. Funding for open access charge: ANR contract.
Conflict of interest statement. None declared.
REFERENCES
- 1. Shultz J.M., Espinel Z., Espinola M., Rechkemmer A.. Distinguishing epidemiological features of the 2013–2016 West Africa Ebola virus disease outbreak. Disaster Health. 2016; 3:78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kuhn J.H., Becker S., Ebihara H., Geisbert T.W., Johnson K.M., Kawaoka Y., Lipkin W.I., Negredo A.I., Netesov S.V., Nichol S.T. et al. . Proposal for a revised taxonomy of the family Filoviridae: classification, names of taxa and viruses, and virus abbreviations. Arch. Virol. 2010; 155:2083–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barrette R.W., Xu L., Rowland J.M., McIntosh M.T.. Current perspectives on the phylogeny of Filoviridae. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2011; 11:1514–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Afonso C.L., Amarasinghe G.K., Bányai K., Bào Y., Basler C.F., Bavari S., Bejerman N., Blasdell K.R., Briand F.X., Briese T., Bukreyev A. et al. . Taxonomy of the order Mononegavirales: update 2016. Arch. Virol. 2016; 161:2351–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sanchez A., Kiley M.P., Holloway B.P., Auperin D.D.. Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res. 1993; 29:215–240. [DOI] [PubMed] [Google Scholar]
- 6. Elliott L.H., Sanchez A., Holloway B.P., Kiley M.P., McCormick J.B.. Ebola protein analyses for the determination of genetic organization. Arch. Virol. 1993; 133:423–436. [DOI] [PubMed] [Google Scholar]
- 7. Nanbo A., Watanabe S., Halfmann P., Kawaoka Y.. The spatio-temporal distribution dynamics of Ebola virus proteins and RNA in infected cells. Sci. Rep. 2013; 3:1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martin B., Canard B., Decroly E.. Filovirus proteins for antiviral drug discovery: structure/function bases of the replication cycle. Antiviral Res. 2017; 141:48–61. [DOI] [PubMed] [Google Scholar]
- 9. Poch O., Blumberg B.M., Bougueleret L., Tordo N.. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J. Gen. Virol. 1990; 71:1153–1162. [DOI] [PubMed] [Google Scholar]
- 10. Liang B., Li Z., Jenni S., Rahmeh A.A., Morin B., Grant T., Grigorieff N., Harrison S.C., Whelan SPJ.. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell. 2015; 162:314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morin B., Liang B., Gardner E., Ross R.A., Whelan S.P.J.. An in vitro RNA synthesis assay for rabies virus defines ribonucleoprotein interactions critical for polymerase activity. J. Virol. 2017; 91:e01508-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abraham G., Rhodes D.P., Banerjee A.K.. The 5′ terminal structure of the methylated mRNA synthesized in vitro by vesicular stomatitis virus. Cell. 1975; 5:51–58. [DOI] [PubMed] [Google Scholar]
- 13. Abraham G., Rhodes D.P., Banerjee A.K.. Novel initiation of RNA synthesis in vitro by vesicular stomatitis virus. Nature. 1975; 255:37–40. [DOI] [PubMed] [Google Scholar]
- 14. Gupta K.C., Roy P.. Alternate capping mechanisms for transcription of spring viremia of carp virus: evidence for independent mRNA initiation. J. Virol. 1980; 33:292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barik S. The structure of the 5′ terminal cap of the respiratory syncytial virus mRNA. J. Gen. Virol. 1993; 74:485–490. [DOI] [PubMed] [Google Scholar]
- 16. Ogino T., Banerjee A.K.. The HR motif in the RNA-dependent RNA polymerase L protein of Chandipura virus is required for unconventional mRNA-capping activity. J. Gen. Virol. 2010; 91:1311–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ogino T., Banerjee A.K.. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol. Cell. 2007; 25:85–97. [DOI] [PubMed] [Google Scholar]
- 18. Tekes G., Rahmeh A.A., Whelan S.P.J.. A freeze frame view of vesicular stomatitis virus transcription defines a minimal length of RNA for 5′ processing. PLoS Pathog. 2011; 7:e1002073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ferron F., Longhi S., Henrissat B., Canard B.. Viral RNA-polymerases – a predicted 2′-O-ribose methyltransferase domain shared by all Mononegavirales. Trends Biochem. Sci. 2002; 27:222–224. [DOI] [PubMed] [Google Scholar]
- 20. Bujnicki J.M., Rychlewski L.. In silico identification, structure prediction and phylogenetic analysis of the 2′-O-ribose (cap 1) methyltransferase domain in the large structural protein of ssRNA negative-strand viruses. Protein Eng. 2002; 15:101–108. [DOI] [PubMed] [Google Scholar]
- 21. Li J., Fontaine-Rodriguez E.C., Whelan S.P.J.. Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J. Virol. 2005; 79:13373–13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rahmeh A.A., Li J., Kranzusch P.J., Whelan S.P.J.. Ribose 2′-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J. Virol. 2009; 83:11043–11050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Testa D., Banerjee A.K.. Two methyltransferase activities in the purified virions of vesicular stomatitis virus. J. Virol. 1977; 24:786–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shatkin A.J. Capping of eucaryotic mRNAs. Cell. 1976; 9:645–653. [DOI] [PubMed] [Google Scholar]
- 25. Decroly E., Ferron F., Lescar J., Canard B.. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 2011; 10:51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paesen G.C., Collet A., Sallamand C., Debart F., Vasseur J.J., Canard B., Decroly E., Grimes J.M.. X-ray structure and activities of an essential Mononegavirales L-protein domain. Nat. Commun. 2015; 6:8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peyrane F., Selisko B., Decroly E., Vasseur J.J., Benarroch D., Canard B., Alvarez K.. High-yield production of short GpppA- and 7MeGpppA-capped RNAs and HPLC-monitoring of methyltransfer reactions at the guanine-N7 and adenosine-2’O positions. Nucleic Acids Res. 2007; 35:e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barral K., Sallamand C., Petzold C., Coutard B., Collet A., Thillier Y., Zimmermann J., Vasseur J.J., Canard B., Rohayem J., Debart F. et al. . Development of specific dengue virus 2′-O- and N7-methyltransferase assays for antiviral drug screening. Antiviral Res. 2013; 99:292–300. [DOI] [PubMed] [Google Scholar]
- 29. Coutard B., Barral K., Lichière J., Selisko B., Martin B., Aouadi W., Lombardia M.O., Debart F., Vasseur J.J., Guillemot J.C. et al. . Zika virus Methyltransferase: Structure and functions for drug design perspectives. J. Virol. 2017; 91:e02202-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lavergne T., Bertrand J.R., Vasseur J.J., Debart F.. A base-labile group for 2′-OH protection of ribonucleosides: a major challenge for RNA synthesis. Chemistry. 14:9135–9138. [DOI] [PubMed] [Google Scholar]
- 31. Lavergne T., Janin M., Dupouy C., Vasseur J.J., Debart F.. Chemical synthesis of RNA with base-labile 2′-o-(pivaloyloxymethyl)-protected ribonucleoside phosphoramidites. Curr. Protoc. Nucleic Acid Chem. 2010; doi:10.1002/0471142700.nc0319s43. [DOI] [PubMed] [Google Scholar]
- 32. Zlatev I., Lavergne T., Debart F., Vasseur J.J., Manoharan M., Morvan F.. Efficient solid-phase chemical synthesis of 5′-triphosphates of DNA, RNA, and their analogues. Org. Lett. 2010; 12:2190–2193. [DOI] [PubMed] [Google Scholar]
- 33. Thillier Y., Decroly E., Morvan F., Canard B., Vasseur J.-J., Debart F.. Synthesis of 5′ cap-0 and cap-1 RNAs using solid-phase chemistry coupled with enzymatic methylation by human (guanine-N7)-methyltransferase. RNA. 2012; 18:856–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dong H., Chang D.C., Hua M.H., Lim S.P., Chionh Y.H., Hia F., Lee Y.H., Kukkaro P., Lok S.M., Dedon P.C. et al. . 2′-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog. 2012; 8:e1002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ogino T., Kobayashi M., Iwama M., Mizumoto K.. Sendai virus RNA-dependent RNA polymerase L protein catalyzes cap methylation of virus-specific mRNA. J. Biol. Chem. 2005; 280:4429–4435. [DOI] [PubMed] [Google Scholar]
- 36. Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E.. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010; 6:e1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G. et al. . Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011; 7:e1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dong H., Ray D., Ren S., Zhang B., Puig-Basagoiti F., Takagi Y., Ho C.K., Li H., Shi PY.. Distinct RNA elements confer specificity to flavivirus RNA cap methylation events. J. Virol. 2007; 81:4412–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chung K.Y., Dong H., Chao A.T., Shi P.Y., Lescar J., Lim S.P.. Higher catalytic efficiency of N-7-methylation is responsible for processive N-7 and 2′-O methyltransferase activity in dengue virus. Virology. 2010; 402:52–60. [DOI] [PubMed] [Google Scholar]
- 40. Barral K., Sallamand C., Petzold C., Coutard B., Collet A., Thillier Y., Zimmermann J., Vasseur J.J., Canard B., Rohayem J. et al. . Development of specific dengue virus 2′-O- and N7-methyltransferase assays for antiviral drug screening. Antiviral Res. 2013; 99:292–300. [DOI] [PubMed] [Google Scholar]
- 41. Hu G., Oguro A., Li C., Gershon P.D., Quiocho F.A.. The ‘cap-binding slot’ of an mRNA cap-binding protein: quantitative effects of aromatic side chain choice in the double-stacking sandwich with cap. Biochemistry (Mosc.). 2002; 41:7677–7687. [DOI] [PubMed] [Google Scholar]
- 42. Egloff M.-P., Decroly E., Malet H., Selisko B., Benarroch D., Ferron F., Canard B.. Structural and functional analysis of methylation and 5′-RNA sequence requirements of short capped RNAs by the methyltransferase domain of dengue virus NS5. J. Mol. Biol. 2007; 372:723–736. [DOI] [PubMed] [Google Scholar]
- 43. Zhou Y., Ray D., Zhao Y., Dong H., Ren S., Li Z., Guo Y., Bernard K.A., Shi P.Y., Li H.. Structure and function of flavivirus NS5 methyltransferase. J. Virol. 2007; 81:3891–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dong H., Chang D.C., Xie X., Toh Y.X., Chung K.Y., Zou G., Lescar J., Lim S.P., Shi P.Y.. Biochemical and genetic characterization of dengue virus methyltransferase. Virology. 2010; 405:568–578. [DOI] [PubMed] [Google Scholar]
- 45. Ray D., Shah A., Tilgner M., Guo Y., Zhao Y., Dong H., Deas T.S., Zhou Y., Li H., Shi P.Y.. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 2006; 80:8362–8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Egloff M.-P., Benarroch D., Selisko B., Romette J.-L., Canard B.. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002; 21:2757–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fabrega C., Hausmann S., Shen V., Shuman S., Lima C.D.. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol. Cell. 2004; 13:77–89. [DOI] [PubMed] [Google Scholar]
- 48. Hodel A.E., Gershon P.D., Quiocho F.A.. Structural basis for sequence-nonspecific recognition of 5′-capped mRNA by a cap-modifying enzyme. Mol. Cell. 1998; 1:443–447. [DOI] [PubMed] [Google Scholar]
- 49. Hager J., Staker B.L., Bugl H., Jakob U.. Active site in RrmJ, a heat shock-induced methyltransferase. J. Biol. Chem. 2002; 277:41978–41986. [DOI] [PubMed] [Google Scholar]
- 50. Decroly E., Imbert I., Coutard B., Bouvet M., Selisko B., Alvarez K., Gorbalenya A.E., Snijder E.J., Canard B.. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2’O)-methyltransferase activity. J. Virol. 2008; 82:8071–8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Aouadi W., Blanjoie A., Vasseur J.J., Debart F., Canard B., Decroly E.. Binding of the methyl donor S-Adenosyl-l-Methionine to middle east respiratory syndrome coronavirus 2′-O-Methyltransferase nsp16 promotes recruitment of the allosteric activator nsp10. J. Virol. 2017; 91:e02217-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McIntyre W., Netzband R., Bonenfant G., Biegel J.M., Miller C., Fuchs G., Henderson E., Arra M., Canki M., Fabris D. et al. . Positive-sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Res. 2018; 46:5776–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yuan Z., Guo W., Yang J., Li L., Wang M., Lei Y., Wan Y., Zhao X., Luo N., Cheng P. et al. . PNAS-4, an early DNA damage response gene, induces S phase arrest and apoptosis by activating checkpoint kinases in lung cancer cells. J. Biol. Chem. 2015; 290:14927–14944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chang K.Y., Varani G.. Nucleic acids structure and recognition. Nat. Struct. Biol. 1997; 4(Suppl.):854–858. [PubMed] [Google Scholar]
- 55. Tawar R.G., Duquerroy S., Vonrhein C., Varela P.F., Damier-Piolle L., Castagné N., MacLellan K., Bedouelle H., Bricogne G., Bhella D. et al. . Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science. 2009; 326:1279–1283. [DOI] [PubMed] [Google Scholar]
- 56. Karikó K., Buckstein M., Ni H., Weissman D.. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005; 23:165–175. [DOI] [PubMed] [Google Scholar]
- 57. Züst R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S. et al. . Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011; 12:137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schuberth-Wagner C., Ludwig J., Bruder A.K., Herzner A.M., Zillinger T., Goldeck M., Schmidt T., Schmid-Burgk J.L., Kerber R., Wolter S. et al. . A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N1-2’O-Methylated self RNA. Immunity. 2015; 43:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Devarkar S.C., Wang C., Miller M.T., Ramanathan A., Jiang F., Khan A.G., Patel S.S., Marcotrigiano J.. Structural basis for m7G recognition and 2′-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.Y., Schneller S., Zust R., Dong H. et al. . 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010; 468:452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kumar P., Sweeney T.R., Skabkin M.A., Skabkina O.V., Hellen C.U., Pestova T.V.. Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′-terminal regions of cap0-, cap1- and 5′ppp- mRNAs. Nucleic Acids Res. 2014; 42:3228–3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shabman R.S., Jabado O.J., Mire C.E., Stockwell T.B., Edwards M., Mahajan M., Geisbert T.W., Basler C.F. et al. . Deep sequencing identifies noncanonical editing of Ebola and Marburg virus RNAs in infected cells. mBio. 2014; 5:e02011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Brody T., Yavatkar A.S., Park D.S., Kuzin A., Ross J., Odenwald W.F.. Flavivirus and Filovirus EvoPrinters: New alignment tools for the comparative analysis of viral evolution. PLoS Negl. Trop. Dis. 2017; 11:e0005673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yi-Brunozzi H.Y., Easterwood L.M., Kamilar G.M., Beal P.A.. Synthetic substrate analogs for the RNA-editing adenosine deaminase ADAR-2. Nucleic Acids Res. 1999; 27:2912–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tao W., Gan T., Guo M., Xu Y., Zhong J.. Novel stable ebola virus minigenome replicon reveals remarkable stability of the viral genome. J. Virol. 2017; 91:e01316-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Reid S.P., Valmas C., Martinez O., Sanchez F.M., Basler C.F.. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J. Virol. 2007; 81:13469–13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xu W., Edwards M.R., Borek D.M., Feagins A.R., Mittal A., Alinger J.B., Berry K.N., Yen B., Hamilton J., Brett T.J. et al. . Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1. Cell Host Microbe. 2014; 16:187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.