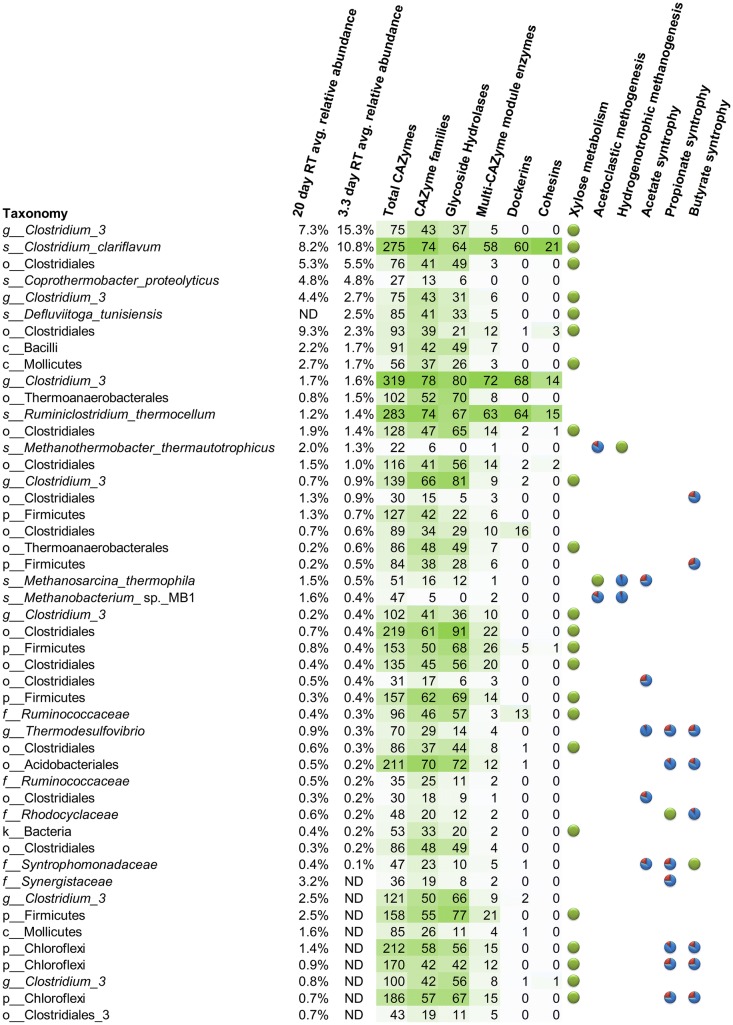
Fig. 5.
Switchgrass digester microbial community with CAZymes profiles and key metabolic pathways. Each microbe listed was found in at least four of fifteen metagenome samples taken from three reactors operating at residence times (RT) of 20, 10, 5, and 3.3 days. Taxonomy was assigned using CheckM; species were manually assigned if its average nucleotide identity was greater than 95% the type strain genome. Microbes were sorted by average relative abundance at the 3.3-day RT. Average relative abundance was not determined (ND) for microbes whose metagenome bins were less than 40% complete or had greater than 20% contamination. Darker green colors indicate higher counts of CAZyme markers. Marker sets used to evaluate metabolic potential were taken from MetaCyc.org and Worm et al. [72]. Complete marker sets are indicated by green pie graphs, while incomplete marker sets with greater than 70% completeness are indicated by red and blue pie graphs, where red indicates the percentage of missing markers