Abstract
The aberrant expression of microRNA (miRNA)-199a-5p has been frequently reported in a number of cancer types, but to the best of our knowledge, this has not been reported in ovarian cancer (OC). The role and the molecular mechanism of miR-199a-5p in OC have not been reported. Therefore, the present study investigated the effects of miR-199a-5p overexpression on the proliferation and invasion of OC cells. The level of miR-199a-5p in OC cell lines was determined by reverse transcription-quantitative polymerase chain reaction. The miR-199a-5p mimic was transiently transfected into OC cells using Lipofectamine™ 2000 reagent. Subsequently, the BrdU-ELISA results indicated that the exogenous expression of miR-199a-5p inhibited cell proliferation. In addition, miR-199a-5p overexpression was able to inhibit the invasion of HO-8910 and ES-2 cells. RT-qPCR was performed to determine the expression of matrix metalloproteinase (MMP)-2 and −9 in OC cells. NF-κB1 expression was reduced by upregulation of miR-199a-5p. Bioinformatics analysis predicted that NF-κB1 was a potential target of miR-199a-5p. Luciferase reporter assay further confirmed that miR-199a-5p was able to directly target the 3′UTR of NF-κB1. In conclusion, miRNA-199a-5p may suppress the proliferation and invasion of human ovarian cancer cells by directly targeting NF-κB1.
Keywords: microRNA-199a-5p, ovarian cancer, proliferation, invasion
Introduction
Ovarian cancer results in abnormal cells that have the ability to invade or spread to other part of the body. Recently, numerous studies confirmed that several genes are closely associated with human ovarian cancer (1,2). However, the precise molecular mechanisms of ovarian cancer remain to be clarified. To date, studies on mechanisms of OC have focused on known genes (3–7), but microRNAs (miRNAs) might also provide further insights into OC.
The discovery of non-coding RNA in the human genome was an important conceptual breakthrough in the post-genomic sequencing era (8–12). miRNAs constitute a class of small, non-coding RNA molecules with a length of 19-22, that modulates gene expression. As a new layer of gene-regulation mechanism, regulation by miRNAs is achieved through imperfect pairing with target mRNAs of protein-coding genes at the transcriptional or post-transcriptional level (13). Furthermore, miRNAs can regulate proliferation, differentiation and apoptosis as well as cancer initiation and progression (14). A number of studies indicated that miRNAs were differentially regulated in various tumors, including colorectal (15), breast (16,17) and lung cancer (18,19).
Recently, a number of reports show that the majority of miRNAs are aberrantly expressed in multiple types of cancer, including OC. For example, miR-17-5p was overexpressed in OC cells, and the upregulation of miR-17-5p significantly promoted the proliferation, migration, invasion and epithelial to mesenchymal transition (EMT) of OC cells by targeting phosphatase and tensin homolog signaling (20). Niu et al (21) demonstrated that miR-205 expression was associated with the motility of OC cells by targeting zinc finger E-Box binding homeobox 1. miR-661 promoted the proliferation of OC cells by directly targeting inositol polyphosphate-5-phosphatase J (22). These three miRNAs (miR-17-5p, miR-205 and miR-661) act as oncogenes; some miRNAs with tumor suppressor functions were also studied in OC. For example, the overexpression of miR-30d reversed transforming growth factor-β1-induced EMT of OC cells by decreasing snail expression (23). The level of miR-127 was markedly downregulated in OC tissues, and the overexpression of miR-127 inhibited the proliferation of OC cells by targeting cyclin G1 (24). miR-302a was also demonstrated to be involved in the suppression of syndecan 1 to inhibit the tumorigenicity of OC cells (25).
miR-199a-5p has been indicated to be involved in the regulation of multiple biological processes, including cell proliferation, apoptosis, migration and invasion (26–31). In recent years, miR-199a-5p was considered as a tumor suppressor and downregulated in prostate, gastric and non-small cell lung cancer (31–33). Guo et al (27) reported that miR-199a-5p functioned as a tumor suppressor by inhibiting proliferation and inducing apoptosis in liver cancer cells by targeting hexokinase 2.
In the present study, the downregulation of miR-199a-5p was observed in HO-8910 and ES-2 OC cell lines. The upregulation of miR-199a-5p was able to inhibit the proliferation and invasion of OC cells. Furthermore, NF-κB1 was identified as a potential target of miR-199a-5p in OC, and miR-199a-5p may function as a tumor suppressor by downregulating NF-κB1. Therefore, these results reveal critical roles for miR-199a-5p in the pathogenesis of OC and miR-199a-5p may be a potential target for the treatment of ovarian tumors.
Materials and methods
Cell culture and miRNA transfection
Human OC cell lines, HO-8910 and ES-2, and an immortalized normal human fallopian tube epithelial cell line, FTE187, were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). All cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) 100 U/ml penicillin and 100 µg/ml streptomycin (Lonza Group, Ltd., Basel, Switzerland). The cells were cultured in 0.1% gelatin-coated culture flasks at 37°C with a humidified atmosphere of 5% CO2. To increase the expression of miR-199a-5p in cell lines, 50 nM miR-199a-5p mimic (sequence: 5′-cccaguguucagacuaccuguuc-3′; RiboBio Co., Ltd., Guangzhou, China) was transfected into HO-8910 and ES-2 cells. miR-negative control (sequence: 5′-catttacgcaguguucagacuaccuguuc-3′; RiboBio Co., Ltd.) was also transfected into HO-8910 and ES-2 cells, respectively. A total of 24 h prior to transfection, when the cells were at ~40-60% confluence, the cell medium was changed to antibiotic-free medium. After 24 h, the cells were transfected with 50 nM miR-199a-5p mimic and miR-negative control using Lipofectamine™ 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The expression of miR-199a-5p and four genes (E-cadherin, vimentin, MMP-2 and MMP-9) was analyzed with SYBR Green II (Takara Co., Ltd. Dalian China) and a RT-qPCR system (MJ Research; Bio-Rad Laboratories, Inc., Hercules, CA, USA). Briefly, the total RNA from HO-8910 and ES-2 cells was extracted using Trizol reagent (Thermo Fisher Scientific, Inc.). A total of 2 mg RNA was used for gene-specific RT-qPCR using a one-step RT-PCR kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's instructions. Complementary DNA (cDNA) was subjected to 40 cycles of amplification. The sequences of the primers are as follows: miR-199a-5p forward, 5′-CCGGGATCCGCAAACTCAGCTTTAC-3′ and reverse 5′-CGGAATTCGTGGCGACCGTGATACC-3′; E-cadherin forward, 5′-TACACTGCCCAGGAGCCAGA-3′ and reverse, 5′-TGGCACCAGTGTCCGGATTA-3′; N-cadherin forward, 5′-CGAATGGATGAAAGACCCATCC-3′ and reverse, 5′-GGAGCCACTGCCTTCATAGTCAA-3′; vimentin, forward, 5′-GCTGAATGACCGCTTCGCCAACT'-3′ and revere, 5′-GCTCCCGCATCTCCTCCTCGTA-3′; MMP-2, forward, 5′-CTGCGGTTTTCTCGAATCCA-3′ and reverse, 5′-GGGTATCCATCGCCATGCT-3′; MMP-9 forward, 5′-CCCTGGAGACCTGAGAACCA-3′ and revere, 5′-CCACCCGAGTGTAACCATAGC-3′; U6, forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; GAPDH forward, 5′-GAGTCAACGGATTTGGTCGTATTG-3′ and reverse, 5′-CCTGGAAGATGGTGATGGGATT-3′. SYBR Green II was used for detection of signal fluorescence and ROX was used as the control fluorescence. Reverse transcription was performed on a GeneAmp PCR system 9700 (Applied Biosystems; Thermo Fisher Scientific, Inc.), and qPCR was performed on ABI 7500 real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). All samples were processed at the same time to avoid inter-experiment variance. The thermocycling conditions were as follows: A holding step at 95°C for 30 sec, and 40 cycles of 95°C for 5 sec and 60°C for 30 sec.
Each experiment was performed in triplicate, and the mean value was used for analysis. The expression level was analyzed by using the ΔΔCq method. ΔCq values represent normalized target genes levels with respect to the internal control. ΔΔCq values were calculated as the ΔCq of each test sample minus the mean ΔCq of the calibrator samples for each target gene. U6 small nuclear RNA and GAPDH mRNA were used for normalization. The fold change was calculated using the method 2−ΔΔCq (34). Each treatment was performed in triplicate in three independent experiments.
Cell proliferation and invasion
To study the role of miR-199a-5p mimic in the proliferation of HO-8910 and ES-2 cells, 5×104 cells were seeded in a 96-well plate and allowed to grow for 24 h in DMEM at 37°C with 10% FBS. The medium was subsequently removed, and the cells were transfected with 50 nM miR-199a-5p mimic or miR-NC for 24 h at 37°C. Cell proliferation 5-bromo-2-deoxyuridine (BrdU)-ELISA colorimetric kit (Roche Diagnostics, Indianapolis, IN, USA) was used to assess cell proliferation according to the manufacturer's instructions. Optical densities were determined at a wavelength of 530 nm using a spectrophotometric plate reader.
Transwell Matrigel invasion assay
The effect of miR-199a-5p on the invasive ability of OC cells was also investigated using Transwell chambers (8-mm pore size; Corning, USA) pre-coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) that included extracellular matrix proteins. In brief, after serum-starvation for 24 h, 2×105 cells were suspended in 100 µl serum-free DMEM, and seeded in the top chamber, and 600 µl DMEM containing 10% FBS was added to the lower chamber. After 24 h incubation at 37°C in a 5% CO2 atmosphere, cells that remained on the upper surface of the membrane were removed by cotton swabs and penetrating cells were fixed in methanol, and then stained with 0.1% crystal violet at 25°C for 12 h. Cell invasion was quantified by counting cells on the lower surface using phase contrast microscopy with ×40 magnification for eight fields of view.
Western blot analysis
For protein extraction, HO-8910 and ES-2 cells were washed twice in cold phosphate-buffered saline and subsequently lysed in radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology, Haimen, China) with a protease inhibitor-PMSF (Beyotime Institute of Biotechnology). The protein concentration of cell lysates was quantified by using the OCA kit (Beyotime Institute of Biotechnology). Equal quantities (50 µg) of total protein were separated by 10% SDS-PAGE, and transferred to a polyvinylidene fluoride membrane (Merck Millipore). The membranes were blocked in 5% skimmed milk diluted with Tris-buffered saline Tween-20 at room temperature for 1 h and incubated overnight at 4°C with primary anti-nuclear factor-κB1 (NF-κB1) antibody (cat. no. 13586, 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA). The membranes were subsequently incubated with a goat anti-rabbit horseradish peroxidase-conjugated immunoglobulin G secondary antibody (cat. no. A0208, 1:2,000; Beyotime Institute of Biotechnology) for 2 h. The proteins were visualized using enhanced chemiluminescence-plus reagents (Beyotime Institute of Biotechnology). The density of the bands was measured using Image J software (version 1.5b; National Institutes of Health USA), and the values were normalized to the densitometric values of GAPDH (1:1,000; Cell Signaling Technology, Inc.) in each sample.
Measurement of MMP-2 and MMP-9 levels
ELISA kits (USCN Life Science, Wuhan, China) was used to determine the levels of MMP-2 and −9 in the cells culture supernatants based on the manufacturer's instructions.
Luciferase reporter assay
NF-κB1 was predicted to be a direct target of miR-199a-5p by using online prediction software RNA22 (https://cm.jefferson.edu/rna22/Interactive/), TargetScan (http://www.targetscan.org/vert_71/) and PITA (https://genie.weizmann.ac.il/pubs/mir07/mir07_data.html). Then, HO-8910 and ES-2 cells (2×106/well) were seeded in 24-well plates and incubated overnight at 37°C prior to transfection. The cells were co-transfected with the 20 ng of NF-κB1-3′ untranslated region (UTR) wild-type (WT) or mutant (MUT) reporter plasmid, 50 nm of miR-199a-5p mimic or miR-negative control (NC) (Promega Corporation, Madison, WI, USA) using Lipofectamine™ 2000. Firefly and Renilla luciferase activities were quantified using a dual luciferase reporter system (Promega Corporation) according to the manufacturer's instructions 48 h following co-transfection. Each treatment was performed in triplicate in three independent experiments.
Statistical analysis
All statistical analyses were performed using SPSS (version 18.0; SPSS, Inc., Chicago, IL, USA). Data from each group are expressed as the mean ± standard error of the mean. One-way analysis of variance was used to compare data sets. Bonferroni test was used to compare the variances at the same time with different means and the Tamhane's T2 test was used to compare variances. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-199a-5p level is decreased in OC cell lines
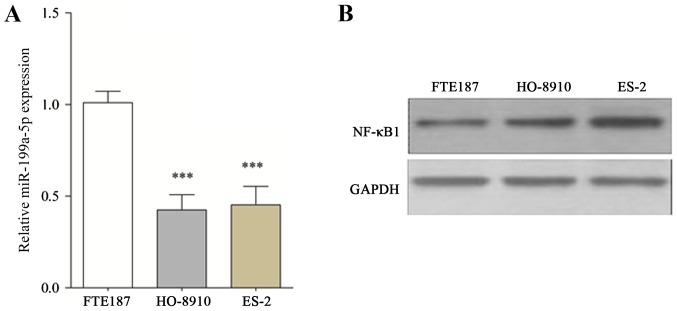
The findings of the present study indicated that miR-199a-5p expression was significantly reduced in HO-8910 and ES-2 cells compared with FTE187 cells (Fig. 1A). Furthermore, NF-κB1 was predicted to be a direct target of miR-199a-5p by using online prediction software RNA22, TargetScan and PITA. The protein expression of NF-κB1, a target of miR-199a-5p, was further assessed in HO-8910 and ES-2 cell lines. The results indicate that NF-κB1 expression in HO-8910 and ES-2 cells was markedly increased compared with FTE187 cells (Fig. 1B).
Figure 1.
Altered expression of miR-199a-5p and NF-κB1 in OC cell lines. (A) The relative level of miR-199a-5p in OC and FTE187 cell lines by reverse transcription-quantitative polymerase chain reaction. (B) Western blotting of NF-κB1 l. All data (n=10) are presented as the mean ± standard error. ***P<0.001 vs. FTE187. NF-κB1, nuclear factor κB1; OC, ovarian cancer; miR, microRNA.
Cell proliferation and invasion are inhibited by miR-199a-5p
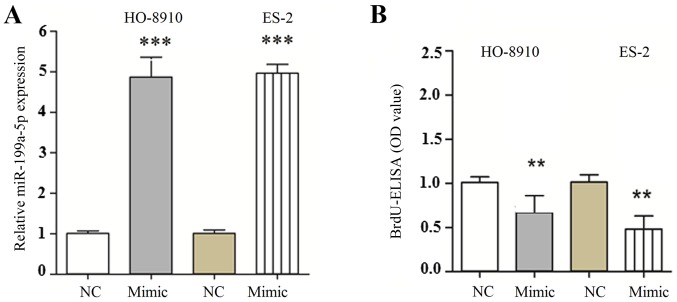
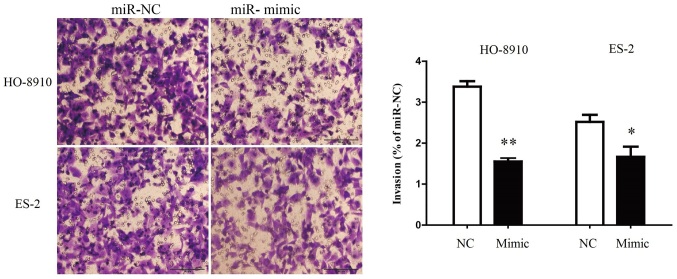
The level of miR-199a-5p mRNA was significantly upregulated following transfection with miR-199a-5p mimic compared with the miR-NC group (Fig. 2A). To investigate the role of miR-199a-5p in the proliferation of OC cells, HO-8910 and ES-2 cell were transfected with miR-199a-5p mimic or miR-NC. Results from the BrdU-ELISA assay indicated that exogenous expression of miR-199a-5p significantly suppressed the proliferation of HO-8910 and ES-2 cells (Fig. 2B). These findings indicated that the upregulation of miR-199a-5p was able to inhibit the proliferation of OC cells. To understand whether the exogenous expression of miR-199a-5p was able to inhibit the invasion of OC cells, miR-199a-5p mimic was transfected into HO-8910 and ES-2 cells and the invasive ability of the cells was evaluated by Transwell invasion assay. It was demonstrated that the number of HO-8910 and ES-2 cells migrating through the Transwell membrane was significantly decreased in the miR-199a-5p mimic group compared with the miR-NC group (Fig. 3). Therefore, these findings indicated that the upregulation of miR-199a-5p suppressed the invasion of OC cells.
Figure 2.
Effects of miR-199a-5p overexpression on the proliferation, apoptosis and cell cycle of HO-8910 and ES-2 cells. HO-8910 and ES-2 cells were transfected with miR-199a-5p mimic or miR-NC. (A) The levels of miR-199a-5p in HO-8910 and ES-2 cells were detected by reverse transcription-quantitative polymerase chain reaction. (B) Cell proliferation was assessed by BrdU-ELISA assay. All data (n=10) are presented as the mean ± standard error. **P<0.01, ***P<0.001 vs. miR-NC. BrdU, 5-bromo-2-deoxyuridine; OD, optical density; miR, microRNA; NC, negative control.
Figure 3.
Overexpression of miR-340 suppresses the invasion of OC cells. HO-8910 and ES-2 cells were transfected with miR-340 mimic and miR-NC, and subsequently seeded into the upper chamber. After 24 h, the HO-8910 and ES-2 cells that migrated through the membrane were stained and quantified with a magnification of ×40 for eight fields of view. Data are expressed as a percentage of the miR-NC group. Data (n=6) are presented as the mean ± standard error. *P<0.05, **P<0.01 vs. miR-NC. miR, microRNA; NC, negative control.
Effect of miR-199a-5p overexpression on MMP-2 and 9 expression in OC cells
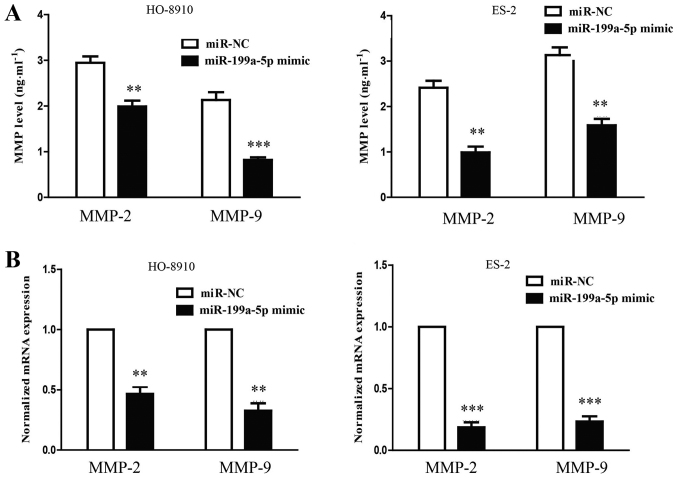
MMPs may be responsible for the impaired invasion of the cells that were transfected with miR-199a-5p mimic. To confirm this hypothesis, the mRNA levels of MMP-2 and 9 were detected by RT-qPCR. The overexpression of miR-199a-5p resulted in significant reductions in the levels of MMP-2 and −9 mRNA (Fig. 4). The results suggested that the upregulation of hsa-miR-199a-5p was able to suppress the invasive ability of HO-8910 and ES-2 cells by potentially downregulating MMP-2 and 9.
Figure 4.
Overexpression of miR-199a-5p decreases the expression and secretion of MMP-2 and MMP-9. The mRNA levels of (A and B) MMP-2 and MMP-9 were examined by reverse transcription-quantitative polymerase chain reaction. All data (n=10) are presented as the mean ± standard error. **P<0.01, ***P<0.001 vs. miR-NC. miR, microRNA; MMP, matrix metallopeptidase; NC, negative control.
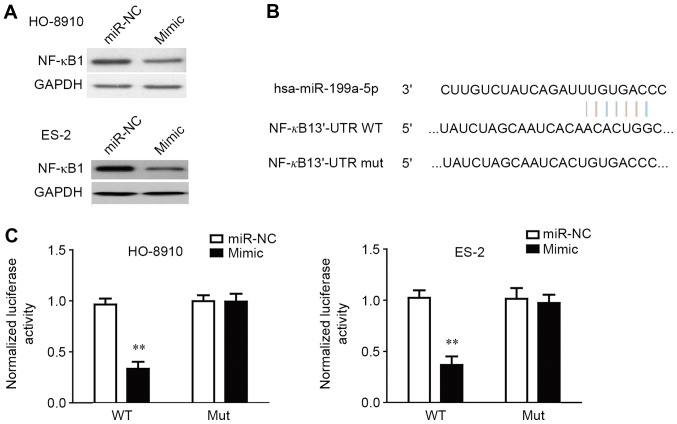
NF-κB1 is a direct target of miR-199a-5p in OC cells
As NF-κB1 was predicted to be a potential direct target of miR-199a-5p by prediction software (RNA22, TargetScan and PITA), the expression of NF-κB1 protein in HO-8910 and ES-2 cells that were transfected with miR-199a-5p mimic was determined by western blotting (Fig. 5A). The results demonstrated that NF-κB1 expression was decreased in the cells that were transfected with miR-199a-5p mimic compared with the cells that were transfected with miR-199a-5p NC (Fig. 5A). To further confirm whether NF-κB1 is a direct target of miR-15a-5p, the 3′-UTR of NF-κB1 was cloned into a luciferase reporter vector, and the putative miR-199a-5p binding site in the NF-κB1 3′-UTR was mutated (Fig. 5B). Luciferase reporter assay was used to determine whether the miR-15a-5p mimic is able to interact with the NF-κB1 3′-UTR. The exogenous expression of the miR-199a-5p mimic was able to significantly suppress the luciferase activity of NF-κB1 3′-UTRWT (Fig. 5C). The effect of miR-199a-5p was abolished by mutating the miR-199a-5p-binding site in the NF-κB1 3′-UTR, which suggested that miR-199a-5p was able to directly negatively regulate NF-κB1.
Figure 5.
NF-κB1 is a direct target of miR-199a-5p. HO-8910 and ES-2 cells were transfected with the miR-199a-5p mimic or miR-NC. A549 and H460 cells were transfected with the miR-199a-5p mimic or miR-NC. (A) The protein expression of NF-κB1 was determined by western blotting, and GAPDH was used as a loading control. (B) Schematic representation of NF-κB1 3′UTRs indicates a putative miRNA target site. (C) Analysis of the relative luciferase activity of OC cells that were co-transfected with NF-κB1-WT or NF-κB1-MUT and miR-NC or miR-199a-5p mimic. All data (n=10) are presented as the mean ± standard error. **P<0.05 vs. miR-NC and NF-κB1-WT co-transfection group. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; miR, microRNA; MUT, mutated; NF-κB1, nuclear factor κB1; NC, negative control; WT, wild-type; UTR, untranslated region.
Discussion
Studies have demonstrated that miRNAs are dysregulated in multiple types of cancer (35–37). Therefore, it is critical for the identification of cancer-specific miRNAs and their targets in order to understand their role in oncogenesis and to identify novel therapeutic targets (38–40). The present study focused on the function of miR-199a-5p in the pathogenesis of OC. Initially, the level of miR-199a-5p was detected in OC cells by RT-qPCR. It was demonstrated that the level of miR-199a-5p was significantly reduced in ovarian cancer cells compared with FTE187 normal human fallopian tube epithelial cells. The results indicated in the present study are consistent with other reports, which have demonstrated that miR-199a-5p was significantly downregulated in colorectal cancer, breast cancer, glioblastoma and oral squamous cell carcinoma (41,42). Therefore, the downregulation of miR-199a-5p is exhibited by multiple types of cancer cells and tissues, indicating that decreased expression of miR-199a-5p may have a critical role in tumorigenesis.
As numerous miRNAs are decreased in various cancer types, it was hypothesized that miRNAs may normally function as tumor suppressor genes (43–45). Therefore, the authors hypothesized that miR-199a-5p was an inhibitory factor of growth in OC cells. As the level of miR-199a-5p was downregulated in cancer cells and tissues, it was hypothesized that the upregulation of miR-199a-5p would lead to cell growth arrest. Using the BrdU-ELISA assay, it was demonstrated that HO-8910 and ES-2 cells that were transfected with the miR-199a-5p mimic exhibited decreased growth compared with the cells transfected with miR-NC.
It has been reported that miR-199a-5p affects the growth, metastasis and apoptosis of cancer cells potentially by targeting cyclin dependent kinase 6, Rho associated coiled-coil containing protein kinase 1 and cMet (30,31,46–49). Although bioinformatic tools might help to reveal the putative mRNA targets of miRNAs, experimental procedures are required for validation. In the resent paper, it was demonstrated that miR-199a-5p was able to target the NF-κB1 miRNA, therefore revealing a potential mechanism associated with ovarian oncogenesis.
As a member of the Rel/NF-κB transcription factor family, NF-κB1 has critical roles in the regulation of immune responses, embryo and cell lineage development, cell-cycle progression, cell apoptosis and tumorigenesis (50–52). The data in the present study indicated that NF-κB1 was a target of miR-199a-5p. First, using western blotting, it was confirmed that the transfection of miR-199a-5p mimic was able to cause a marked decrease in the levels of NF-κB1 protein. In addition, it was indicated that miR-199a-5p was able to directly regulate NF-κB1 expression, as the seed region of miR-199a-5p is able to bind with the 3′ UTR of NF-κB1 mRNA. Furthermore, the luciferase activity of the NF-κB1 3′-UTR was specifically responsive to miR-199a-5p upregulation. Indeed, mutation of the miR-199a-5p binding site abolished the effect of miR-199a-5p on the regulation of luciferase activity.
NF-κB1 facilitates invasion and metastasis of cancer cells partly by transcriptional regulation of MMP-2 and MMP-9 (53). Ferrer-Marin et al (53) have demonstrated that miR-9 suppressed the migration and invasion of uveal melanoma cells partly by targeting NF-κB1 expression and downregulating, MMP-2 and −9, its downstream molecules. An additional study has indicated that miR-9 was able to inhibit the metastasis of melanoma cells by suppressing E-cadherin expression via targeting NF-κB1 (54). In the present study, it was demonstrated that the expression of MMP-2 and MMP-9 was altered in the same pattern as NF-κB1 expression upon the exogenous expression of miR-199a-5p. Therefore, the regulation of MMP-2 and MMP-9 by NF-κB1 may contribute to the invasion and metastasis of OC cells that is mediated by NF-κB1.
In summary, the present study indicated that miR-199a-5p was downregulated in OC cells compared with the normal human fallopian tube epithelial cell line. The transfection of miR-199a-5p inhibited the proliferation and invasion of OC cells by directly targeting NF-κB1. This novel miR-199a-5p/NF-κB1 axis might provide new insights into the molecular mechanisms underlying the progression and metastasis of OC, and the overexpression of miR-199a-5p might be a potential therapeutic strategy for the treatment of OC in the future.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Basic Natural Science Foundation of Xinchang People's Hospital of Zhejiang Province (Zhejiang, China; grant no. 81201101).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XL and BY contributed to experiments and data analyses. ZW contributed to the design of the project and writing of the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zhou X, Hu Y, Dai L, Wang Y, Zhou J, Wang W, Di W, Qiu L. MicroRNA-7 inhibits tumor metastasis and reverses epithelial-mesenchymal transition through AKT/ERK1/2 inactivation by targeting EGFR in epithelial ovarian cancer. PLoS One. 2014;9:e96718. doi: 10.1371/journal.pone.0096718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu T, Hou L, Huang Y. EZH2-specific microRNA-98 inhibits human ovarian cancer stem cell proliferation via regulating the pRb-E2F pathway. Tumour Biol. 2014;35:7239–7247. doi: 10.1007/s13277-014-1950-9. [DOI] [PubMed] [Google Scholar]
- 3.James PA, Sawyer S, Boyle S, Young MA, Kovalenko S, Doherty R, McKinley J, Alsop K, Beshay V, Harris M, et al. Large genomic rearrangements in the familial breast and ovarian cancer gene BRCA1 are associated with an increased frequency of high risk features. Fam Cancer. 2015;14:287–295. doi: 10.1007/s10689-015-9785-0. [DOI] [PubMed] [Google Scholar]
- 4.Koensgen D, Bruennert D, Ungureanu S, Sofroni D, Braicu EI, Sehouli J, Sümnig A, Delogu S, Zygmunt M, Goyal P, et al. Polymorphism of the IL-8 gene and the risk of ovarian cancer. Cytokine. 2015;71:334–338. doi: 10.1016/j.cyto.2014.07.254. [DOI] [PubMed] [Google Scholar]
- 5.Xing BL, Li T, Tang ZH, Jiao L, Ge SM, Qiang X, OuYang J. Cumulative methylation alternations of gene promoters and protein markers for diagnosis of epithelial ovarian cancer. Genet Mol Res. 2015;14:4532–4540. doi: 10.4238/2015.May.4.11. [DOI] [PubMed] [Google Scholar]
- 6.Xu Q, Ding YY, Song LX, Xu JF. Correlation of UGT1A1 and ERCC1 gene polymorphisms with the outcome of combined irinotecan plus cisplatin treatment in recurrent ovarian cancer. Genet Mol Res. 2015;14:7241–7247. doi: 10.4238/2015.June.29.17. [DOI] [PubMed] [Google Scholar]
- 7.Yan H, Tong J, Lin X, Han Q, Huang H. Effect of the WWOX gene on the regulation of the cell cycle and apoptosis in human ovarian cancer stem cells. Mol Med Rep. 2015;12:1783–1788. doi: 10.3892/mmr.2015.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Z, Yu YQ, Ge YZ, Zhu JG, Zhu M, Zhao YC, Xu LW, Yang XB, Geng LG, Dou QL, Jia RP. MicroRNA expression profiles in muscle-invasive bladder cancer: Identification of a four-microRNA signature associated with patient survival. Tumour Biol. 2015;36:8159–8166. doi: 10.1007/s13277-015-3559-z. [DOI] [PubMed] [Google Scholar]
- 9.Itesako T, Seki N, Yoshino H, Chiyomaru T, Yamasaki T, Hidaka H, Yonezawa T, Nohata N, Kinoshita T, Nakagawa M, Enokida H. The microRNA expressions signature of bladder cancer by deep sequencing: The functional significance of the miR-195/497 cluster. PLoS One. 2014;9:e84311. doi: 10.1371/journal.pone.0084311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pignot G, Cizeron-Clairac G, Vacher S, Susini A, Tozlu S, Vieillefond A, Zerbib M, Lidereau R, Debre B, Amsellem-Ouazana D, Bieche I. microRNA expression profile in a large series of bladder tumors: Identification of a 3-miRNA signature associated with aggressiveness of muscle-invasive bladder cancer. Int J Cancer. 2013;132:2479–2491. doi: 10.1002/ijc.27949. [DOI] [PubMed] [Google Scholar]
- 11.Zhang AM, Ma K, Song Y, Wang B, Feng Y, Liu L, Xia X. Genetic polymorphisms of the IFNλ genes are associated with biochemical features in Han Chinese with HCV infection from Yunnan Province, China. Infect Genet Evol. 2014;21:161–165. doi: 10.1016/j.meegid.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 12.Zhang AM, Ma K, Song Y, Feng Y, Duan H, Zhao P, Wang B, Xu G, Li Z, Xia X. Mitochondrial DNAs decreased and correlated with clinical features in HCV patients from Yunnan, China. Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27:2516–2519. doi: 10.3109/19401736.2015.1036255. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Jabbari N, Reavis AN, McDonald JF. Sequence variation among members of the miR-200 microRNA family is correlated with variation in the ability to induce hallmarks of mesenchymal-epithelial transition in ovarian cancer cells. J Ovarian Res. 2014;7:12. doi: 10.1186/1757-2215-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maqbool R, Ismail R, Hussain M. Mutations in MicroRNA genes and they binding sites are infrequently associated with human colorectal cancer in the kashmiri population. Microrna. 2014;2:219–224. doi: 10.2174/2211536602666140102001007. [DOI] [PubMed] [Google Scholar]
- 16.De M, attos-Arruda L, Bottai G, Nuciforo PG, Di Tommaso L, Giovannetti E, Peg V, Losurdo A, Pérez-Garcia J, Masci G, Corsi F, et al. MicroRNA-21 links epithelial-to-mesenchymal transition and inflammatory signals to confer resistance to neoadjuvant trastuzumab and chemotherapy in HER2-positive breast cancer patients. Oncotarget. 2015;6:37269–37280. doi: 10.18632/oncotarget.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Li Y, Qi W, Zhang N, Sun M, Huo Q, Cai C, Lv S, Yang Q. MicroRNA-99a inhibits tumor aggressive phenotypes through regulating HOXA1 in breast cancer cells. Oncotarget. 2015;6:32737–32747. doi: 10.18632/oncotarget.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sestini S, Boeri M, Marchiano A, Pelosi G, Galeone C, Verri C, Suatoni P, Sverzellati N, La Vecchia C, Sozzi G, Pastorino U. Circulating microRNA signature as liquid-biopsy to monitor lung cancer in low-dose computed tomography screening. Oncotarget. 2015;6:32868–32877. doi: 10.18632/oncotarget.5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui R, Kim T, Fassan M, Meng W, Sun HL, Jeon YJ, Vicentini C, Tili E, Peng Y, Scarpa A, et al. MicroRNA-224 is implicated in lung cancer pathogenesis through targeting caspase-3 and caspase-7. Oncotarget. 2015;6:21802–21815. doi: 10.18632/oncotarget.5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang Y, Xu C, Fu Y. MicroRNA-17-5p induces drug resistance and invasion of ovarian carcinoma cells by targeting PTEN signaling. J Biol Res (Thessalon) 2015;22:12. doi: 10.1186/s40709-015-0035-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niu K, Shen W, Zhang Y, Zhao Y, Lu Y. MiR-205 promotes motility of ovarian cancer cells via targeting ZEB1. Gene. 2015;574:330–336. doi: 10.1016/j.gene.2015.08.017. [DOI] [PubMed] [Google Scholar]
- 22.Zhu T, Yuan J, Wang Y, Gong C, Xie Y, Li H. MiR-661 contributed to cell proliferation of human ovarian cancer cells by repressing INPP5J expressions. Biomed Pharmacother. 2015;75:123–128. doi: 10.1016/j.biopha.2015.07.023. [DOI] [PubMed] [Google Scholar]
- 23.Ye Z, Zhao L, Li J, Chen W, Li X. miR-30d blocked transforming growth factor β1-induced epithelial-mesenchymal transition by targeting snail in ovarian cancer cells. Int J Gynecol Cancer. 2015;25:1574–1581. doi: 10.1097/IGC.0000000000000546. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Ma L, Rao Q, Mao Y, Xin Y, Xu H, Li C, Wang X. MiR-1271 inhibits ovarian cancer growth by targeting cyclin G1. Med Sci Monit. 2015;21:3152–3158. doi: 10.12659/MSM.895562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo T, Yu W, Lv S, Zhang C, Tian Y. MiR-302a inhibits the tumorigenicity of ovarian cancer cells by suppression of SDC1. Int J Clin Exp Pathol. 2015;8:4869–4880. [PMC free article] [PubMed] [Google Scholar]
- 26.Mussnich P, Rosa R, Bianco R, Fusco A, D'Angelo D. MiR-199a-5p and miR-375 affect colon cancer cell sensitivity to cetuximab by targeting PHLPP1. Expert Opin Ther Targets. 2015;19:1017–1026. doi: 10.1517/14728222.2015.1057569. [DOI] [PubMed] [Google Scholar]
- 27.Guo W, Qiu Z, Wang Z, Wang Q, Tan N, Chen T, Chen Z, Huang S, Gu J, Li J, et al. MiR-199a-5p is negatively associated with malignancies and regulates glycolysis and lactate production by targeting hexokinase 2 in liver cancer. Hepatology. 2015;62:1132–1144. doi: 10.1002/hep.27929. [DOI] [PubMed] [Google Scholar]
- 28.Kim BK, Yoo HI, Kim I, Park J, Kim Yoon S. FZD6 expression is negatively regulated by miR-199a-5p in human colorectal cancer. BMB Rep. 2015;48:360–366. doi: 10.5483/BMBRep.2015.48.6.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JM, Heo MJ, Lee CG, Yang YM, Kim SG. Increase of miR-199a-5p by protoporphyrin IX, a photocatalyzer, directly inhibits E2F3, sensitizing mesenchymal tumor cells to anti-cancer agents. Oncotarget. 2015;6:3918–3931. doi: 10.18632/oncotarget.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao X, He L, Li T, Lu Y, Miao Y, Liang S, Guo H, Bai M, Xie H, Luo G, et al. SRF expedites metastasis and modulates the epithelial to mesenchymal transition by regulating miR-199a-5p expression in human gastric cancer. Cell Death Differ. 2014;21:1900–1913. doi: 10.1038/cdd.2014.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu Y, Liu J, Jiang B, Chen J, Fu Z, Bai F, Jiang J, Tang Z. MiR-199a-5p loss up-regulated DDR1 aggravated colorectal cancer by activating epithelial-to-mesenchymal transition related signaling. Dig Dis Sci. 2014;59:2163–2172. doi: 10.1007/s10620-014-3136-0. [DOI] [PubMed] [Google Scholar]
- 32.He XJ, Ma YY, Yu S, Jiang XT, Lu YD, Tao L, Wang HP, Hu ZM, Tao HQ. Up-regulated miR-199a-5p in gastric cancer functions as an oncogene and targets klotho. BMC Cancer. 2014;14:218. doi: 10.1186/1471-2407-14-218. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Yi H, Liang B, Jia J, Liang N, Xu H, Ju G, Ma S, Liu X. Differential roles of miR-199a-5p in radiation-induced autophagy in breast cancer cells. FEBS Lett. 2013;587:436–443. doi: 10.1016/j.febslet.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Saito Y, Nakaoka T, Saito H. microRNA-34a as a therapeutic agent against human cancer. J Clin Med. 2015;4:1951–1959. doi: 10.3390/jcm4111951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ono S, Lam S, Nagahara M, Hoon DS. Circulating microRNA Biomarkers as liquid biopsy for cancer patients: Pros and cons of current assays. J Clin Med. 2015;4:1890–1907. doi: 10.3390/jcm4101890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyazaki T, Ikeda K, Sato W, Horie-Inoue K, Okamoto K, Inoue S. MicroRNA library-based functional screening identified androgen-sensitive miR-216a as a player in bicalutamide resistance in prostate cancer. J Clin Med. 2015;4:1853–1865. doi: 10.3390/jcm4101853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.An F, Olaru AV, Mezey E, Xie Q, Li L, Piontek KB, Selaru FM. MicroRNA-224 induces G1/S checkpoint release in liver cancer. J Clin Med. 2015;4:1713–1728. doi: 10.3390/jcm4091713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horsham JL, Kalinowski FC, Epis MR, Ganda C, Brown RA, Leedman PJ. Clinical potential of microRNA-7 in cancer. J Clin Med. 2015;4:1668–1687. doi: 10.3390/jcm4091668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelly BD, Miller N, Sweeney KJ, Durkan GC, Rogers E, Walsh K, Kerin MJ. A Circulating MicroRNA signature as a biomarker for prostate cancer in a high risk group. J Clin Med. 2015;4:1369–1379. doi: 10.3390/jcm4071369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stiuso P, Potenza N, Lombardi A, Ferrandino I, Monaco A, Zappavigna S, Vanacore D, Mosca N, Castiello F, Porto S, et al. MicroRNA-423-5p promotes autophagy in cancer cells and is increased in serum from hepatocarcinoma patients treated with sorafenib. Mol Ther Nucleic Acids. 2015;4:e233. doi: 10.1038/mtna.2015.8. [DOI] [PubMed] [Google Scholar]
- 42.Slattery ML, Mullany LE, Sakoda LC, Samowitz WS, Wolff RK, Stevens JR, Herrick JS. Expression of Wnt-signaling pathway genes and their associations with miRNAs in colorectal cancer. Oncotarget. 2017;9:6075–6085. doi: 10.18632/oncotarget.23636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nunez Lopez YO, Victoria B, Golusinski P, Golusinski W, Masternak MM. Characteristic miRNA expression signature and random forest survival analysis identify potential cancer-driving miRNAs in a broad range of head and neck squamous cell carcinoma subtypes. Rep Pract Oncol Radiother. 2018;23:6–20. doi: 10.1016/j.rpor.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deb B, Uddin A, Chakraborty S. miRNAs and ovarian cancer: An overview. J Cell Physiol. 2018;233:3846–3854. doi: 10.1002/jcp.26095. [DOI] [PubMed] [Google Scholar]
- 45.Weiner-Gorzel K, Dempsey E, Milewska M, McGoldrick A, Toh V, Walsh A, Lindsay S, Gubbins L, Cannon A, Sharpe D, et al. Overexpression of the microRNA miR-433 promotes resistance to paclitaxel through the induction of cellular senescence in ovarian cancer cells. Cancer Med. 2015;4:745–758. doi: 10.1002/cam4.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou J, Liu R, Wang Y, Tang J, Tang S, Chen X, Xia K, Xiong W, Xu D, Wang S, et al. miR-199a-5p regulates the expression of metastasis-associated genes in B16F10 melanoma cells. Int J Clin Exp Pathol. 2014;7:7182–7190. [PMC free article] [PubMed] [Google Scholar]
- 47.Lin HS, Gong JN, Su R, Chen MT, Song L, Shen C, Wang F, Ma YN, Zhao HL, Yu J, et al. miR-199a-5p inhibits monocyte/macrophage differentiation by targeting the activin A type 1B receptor gene and finally reducing C/EBPα expression. J Leukoc Biol. 2014;96:1023–1035. doi: 10.1189/jlb.1A0514-240R. [DOI] [PubMed] [Google Scholar]
- 48.Raimondi L, Amodio N, Di Martino MT, Altomare E, Leotta M, Caracciolo D, Gullà A, Neri A, Taverna S, D'Aquila P, et al. Targeting of multiple myeloma-related angiogenesis by miR-199a-5p mimics: In vitro and in vivo anti-tumor activity. Oncotarget. 2014;5:3039–3054. doi: 10.18632/oncotarget.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu ZY, Lu L, Liang J, Guo XR, Zhang PH, Luo SJ. Keloid microRNA expression analysis and the influence of miR-199a-5p on the proliferation of keloid fibroblasts. Genet Mol Res. 2014;13:2727–2738. doi: 10.4238/2014.April.14.2. [DOI] [PubMed] [Google Scholar]
- 50.Oltulu YM, Coskunpinar E, Ozkan G, Aynaci E, Yildiz P, Isbir T, Yaylim I. Investigation of NF-κB1 and NF-κBIA gene polymorphism in non-small cell lung cancer. Biomed Res Int. 2014;2014:530381. doi: 10.1155/2014/530381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang H, Ma L, Li J, Yu Y, Zhang D, Wei J, Jin H, Xu D, Gao J, Huang C. NF-κB1 inhibits c-Myc protein degradation through suppression of FBW7 expression. Oncotarget. 2014;5:493–505. doi: 10.18632/oncotarget.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu Y, Zhang D, Huang H, Li J, Zhang M, Wan Y, Gao J, Huang C. NF-κB1 p50 promotes p53 protein translation through miR-190 downregulation of PHLPP1. Oncogene. 2014;33:996–1005. doi: 10.1038/onc.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferrer-Marin F, Gutti R, Liu ZJ, Sola-Visner M. MiR-9 contributes to the developmental differences in CXCR-4 expression in human megakaryocytes. J Thromb Haemost. 2014;12:282–285. doi: 10.1111/jth.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen P, Price C, Li Z, Li Y, Cao D, Wiley A, He C, Gurbuxani S, Kunjamma RB, Huang H, et al. miR-9 is an essential oncogenic microRNA specifically overexpressed in mixed lineage leukemia-rearranged leukemia. Proc Natl Acad Sci USA. 2013;110:11511–11516. doi: 10.1073/pnas.1310144110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.