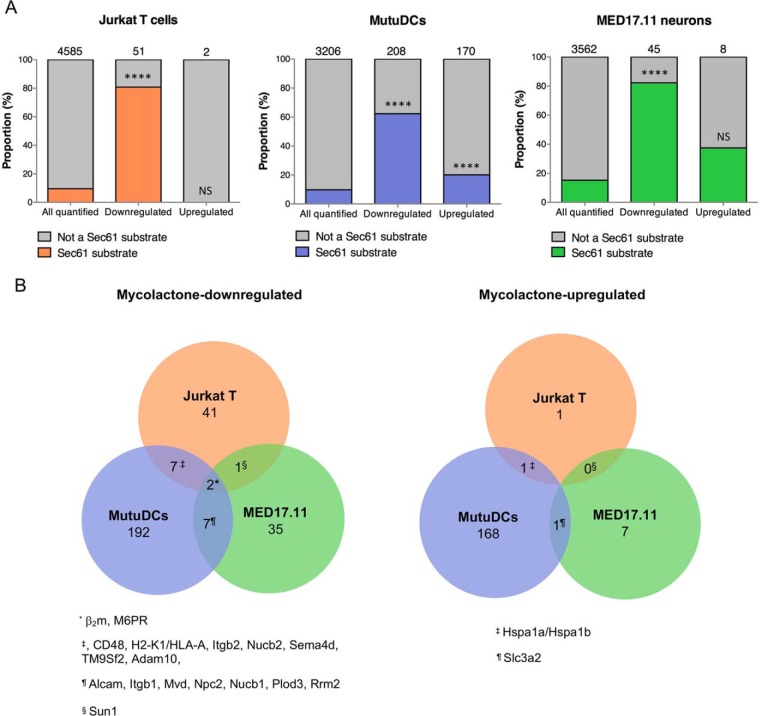
Fig. 1.
Conserved and variable features of mycolactone-induced proteomic alterations. A, The proportion of Sec61 substrates in “all quantified” proteins is compared with that in “mycolactone downregulated” or “mycolactone-upregulated” proteins, in each cell type studied. Jurkat T cells (left), MutuDCs (middle) and MED17.11 cells (right) were treated with mycolactone or vehicle as control in the conditions outlined in Table II. Number of identified proteins in each subset are indicated on the top of each bar, **** p value < 0.0001, ns: not significant, Fisher exact test. B, Venn diagrams representing the overlap between mycolactone downregulated (left) or mycolactone-upregulated (right) proteins across cell types. Human proteins (Jurkat T cells) were matched to their mouse orthologues (MutuDCs and MED17.11 neurons). Proteins that were found downregulated or upregulated by mycolactone in 2 cell types or more are listed.