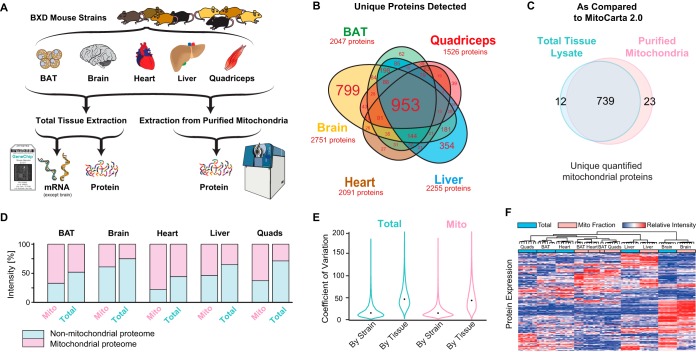
Fig. 1.
Cross-tissue mitochondrial proteomics using SWATH-MS. A, Project design. Eight genetically distinct BXD strains were selected for proteomic analysis in five tissues: BAT, brain, heart, liver, and quadriceps. Each tissue sample was measured for both whole-cell proteome and purified mitoproteome. mRNA was previously measured in the same individuals for all tissues except brain. B, Venn diagram showing the identification of proteins in each tissue. Brain has the most uniquely identified proteins. Font size corresponds to the number of proteins per category. C, Only a few additional mitoproteins are identified in purified mitochondria. This Venn diagram is collation of all tissues' data. D, Total proteome intensities summed and segregated into mitoproteins or non-mitoproteins based on MitoCarta. E, Coefficients of variation for protein intensities across strains and tissues. The median coefficient of variation within tissue across strains was ∼15%, compared with coefficients of variation of ∼45% within-strain across tissues. F, Hierarchical clustering of all protein expression levels across samples. Clustering (Euclidean distance, black lines at top) was performed using all samples proteins, although only 150 proteins are displayed below (y axis) for brevity and clarity in visualization.