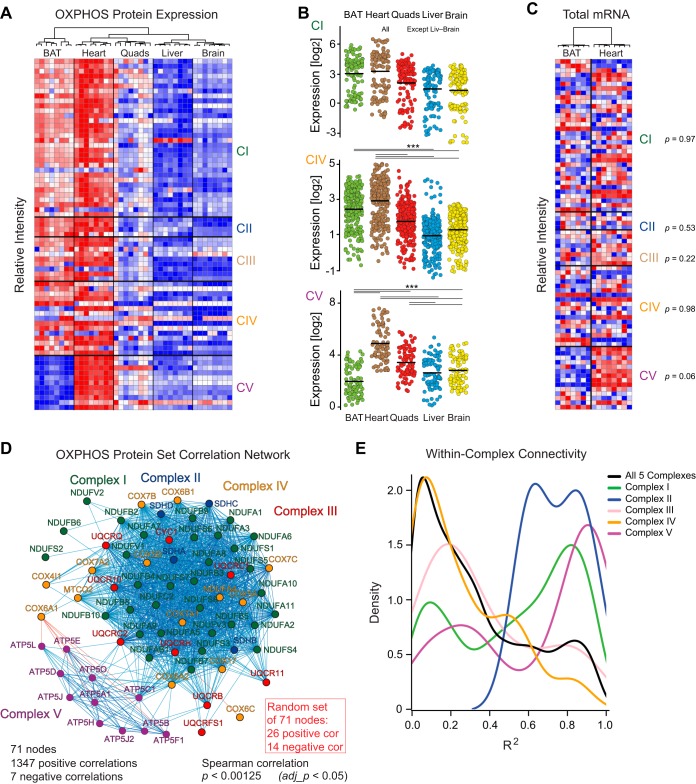
Fig. 3.
Functional protein network based on large-scale proteomics. A, Relative protein intensities of the 71 measured OXPHOS proteins. BAT shows high expression of CI-IV, but the lowest expression of CV proteins (ANOVA post-hoc Dunnett test, p < 0.005 BAT versus all other tissues). B, Significant differences of CIV and CV protein expression across all five tissues (ANOVA with post-hoc Tukey tests, p < 0.005 indicated by ***). For CI, all differences are significant except for liver versus brain (p = 0.11). C, Transcript expression intensities in BAT and heart for the 66 OXPHOS transcripts that also have protein expression data (5 OXPHOS genes have protein measurements but no transcript measurements). Hierarchical clustering separates tissues, yet no complexes have different expression (paired Welch's t-tests, shown at side). D, Covariation network of all five tissues together shows major coexpression of CI-IV whereas CV is distinctly apart because of its contrary regulation in BAT. At adj_p < 0.05, there are 1347 positive correlations and 7 negative correlations between the 71 OXPHOS nodes (proteins). For a set of 71 proteins randomly selected from the same dataset, only 26 positive and 14 negative correlations are observed at the same cutoff. E, Covariation within complexes: Most correlations from CI, CII, and CV consistently correlate within their own complex, whereas CIII and CIV proteins are more variably distributed.