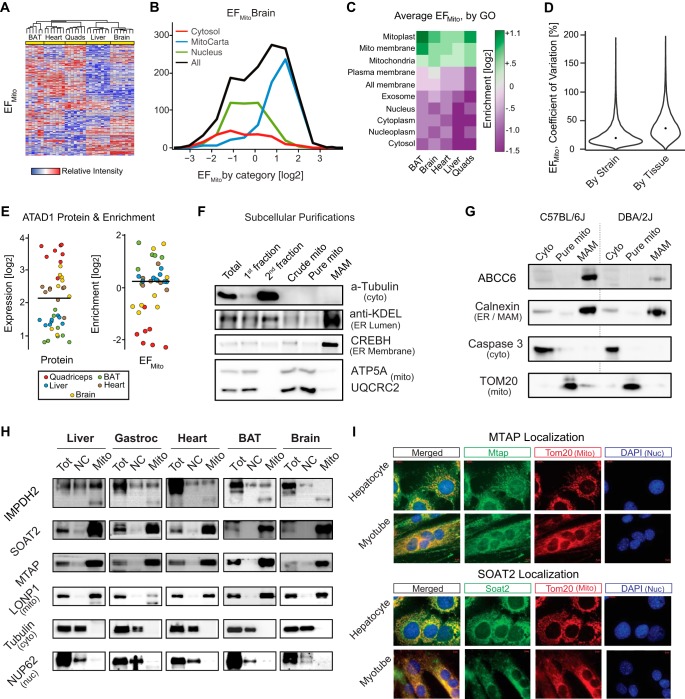
Fig. 4.
Using EFmito to identify mitochondrial localization. A, Cluster analysis of all 3648 proteins based on EFmito. Hierarchical clustering separates all tissues. B, Histogram of EFmito frequencies in brain for gene ontologies of nuclear, cytosolic, or mitochondrial localization. Mitochondrial proteins generally have high EFmito, whereas nuclear and cytosolic proteins generally have low EFmito, though significant overlap is observed. C, Heat map showing the average EFmito across all tissues in a selection of cell compartments, with consistent enrichment of mitoprotein sets. D, Coefficients of variation between strains and tissues for EFmito. Within strains, median is 22%, compared with 39% across tissues, i.e. tissue differences again have a larger impact than do strain differences. E, ATAD1 expression varied among tissues, with quadriceps having the highest expression yet lowest EFmito, suggesting tissue-dependent subcellular localization. F, Fractionations showing the ultracentrifugation purification of the mitochondria from the MAM. G, ABCC6 is consistently localized in the MAM across strains, though trace amounts appear in the cytosol. Abcc6 has a major sequence variant between C57BL/6J and DBA/2J (Williams et al., 2016), which substantially affects expression but not localization. H, Western blots indicate that MTAP, SOAT2, and IMPDH2 are localized in the mitochondria. LONP1, tubulin, and NUP62 were used as mitochondrial, cytosolic and nuclear markers, respectively. NC: Total cell minus the mitochondrial and MAM fractions. I, Mitochondrial and nuclear ICC staining of MTAP and SOAT2 shows the localization of both proteins with mitochondria in hepatocytes and myotubes. Tom20 and DAPI were used to stain mitochondria and nuclei, respectively.