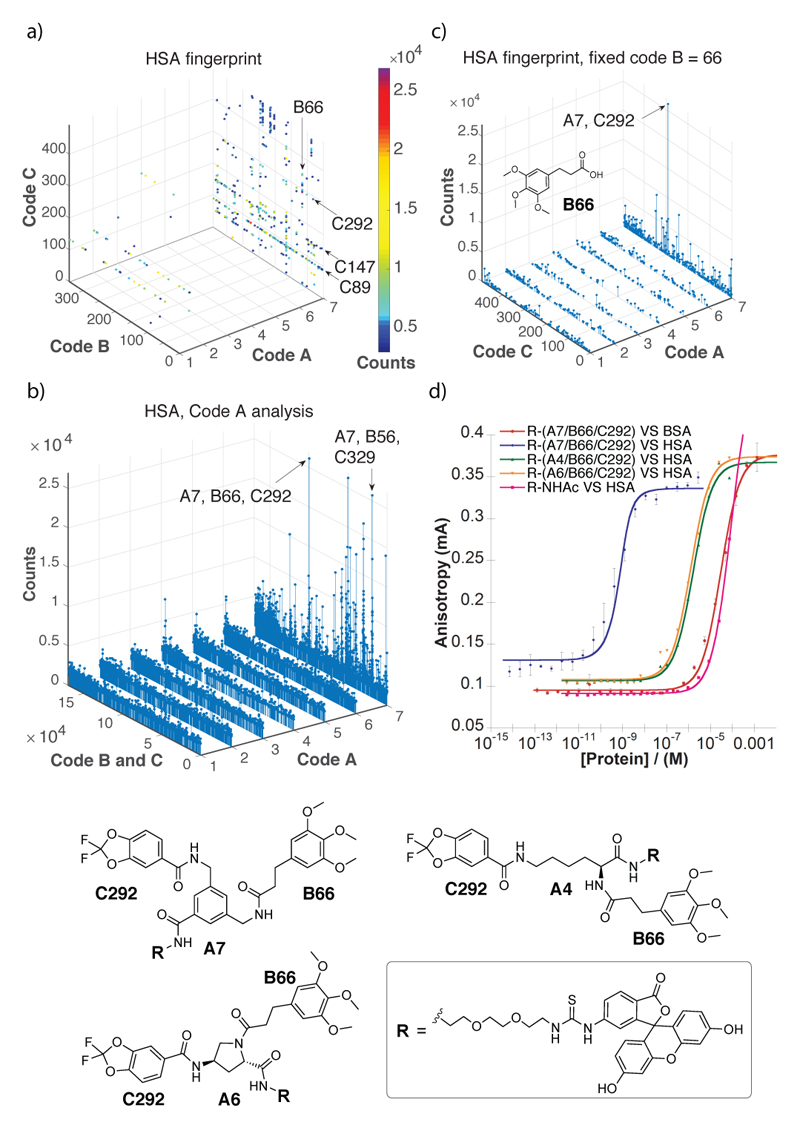
Figure 2.
Results of library selections against human serum albumin (HSA). (a) Selection fingerprint. The individual library members are unambiguously identified by their code A (ranging between 1 and 7), B (ranging between 1 and 343) and C (ranging between 1 and 492). The number of sequence counts for each compound is displayed as spheres of a different colour, with a cut-off threshold set at 3000 counts. In total, 59,596,519 sequence counts were read by high-throughput sequencing. (b) The Code A analysis shows how the different scaffolds (code A, ranging 1 and 7) display different enrichment factors for each building block A and B combination (Code B × Code, ranging between 1 and 1.7×105). (c) Selection fingerprint obtained fixing the code B = 66 (corresponding to 3-(3,4,5-trimethoxyphenyl)propanoic acid) with variable codes A (ranging between 1 and 7) and C (ranging between 1 and 492). This analysis shows the A7/B66/C292 as the most enriched combination of building blocks against HSA. (d) Fluorescence polarization (FP) measurement of FITC-conjugates of the most enriched combination of building blocks (B66/C292), featuring the preferred scaffold A7 (red and blue curves) or scaffolds A4 (green) and A6 (orange). A FITC conjugate of A7/B66/C292 was also tested against bovine serum albumin (BSA) a d revealed a double-digit micromolar dissociation constant (Kd) against that protein (pink curve).