Abstract
Long non-coding RNA (lncRNA) have yet to be linked to cancer metabolism. Here we report that upregulation of the lncRNA LINC00538 (YIYA) promotes glycolysis, cell proliferation and tumor growth in in breast cancer. YIYA associated with the cytosolic cyclin-dependent kinase CDK6 and regulated CDK6-dependent phosphorylation of the fructose bisphosphatase PFK2 (PFKFB3) in a cell cycle-independent manner. In breast cancer cells, these events promoted catalysis of glucose 6-phosphate to fructose-2,6-bisphosphate/fructose-1,6-bisphosphate. CRISPR/Cas9-mediated deletion of YIYA or CDK6 silencing impaired glycolysis and tumor growth in vivo. In clinical specimens of breast cancer, YIYA was expressed in ~40% of cases where it correlated with CDK6 expression and unfavorable survival outcomes. Our results define a functional role for lncRNA in metabolic reprogramming in cancer, with potential clinical implications for its therapeutic targeting.
Keywords: long non-coding RNA, Glucose metabolism, Glycolysis, CDK6, PFKFB3, Fructose Bisphosphatase 2, PFKFB3
Precis
Findings offer a first glimpse into how a long-coding RNA influences cancer metabolism to drive tumor growth.
INTRODUCTION
It has been found that numerous long non-coding RNA transcripts are transcribed in the human genome [1], and play critical roles in diverse cellular processes and diseases, including neurodegenerative diseases, autoimmune diseases, and cancer [2–4]. A long non-coding RNA, named YIYA, was originally demonstrated to be upregulated in multiple cancer types. YIYA is located on 1q41, which is a cancer susceptibility locus [5]. The YIYA gene is also amplified, and its expression is linked to cell cycle regulation and cell proliferation [5].
One of the hallmarks of human cancer is the alteration of glucose metabolism. Elevated glycolysis (also known as the Warburg effect) promotes cancer cell growth, angiogenesis and metastasis [6]. One of the key steps of glucose metabolism is the conversion of glucose 6-phosphate (G6P) to fructose-1,6-bisphosphate (F-1,6-BP) or fructose-2,6-bisphosphate (F-2,6-BP) [7, 8], which is mediated by 6-phosphofructo-1-kinase (PFK1) [9] or 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB3), respectively [10]. The enzymatic activity of PFK1 is allosterically activated by F-2,6-BP [11]. Inhibition of PFKFB3 reduces the enzymatic activity of PFK1 and glucose flux, leading to decreased cancer cell growth [12, 13].
In addition to cell cycle regulation, cyclin-dependent kinase 6 (CDK6) has been suggested to modulate other cellular activities [14]. Mice with CDK6 knockout exhibited impaired hematopoietic development [15]. In addition, recent studies have also indicated that CDK6 regulates the pentose pathway [16]. Inhibition of CDK6 by small molecule inhibitors may impair the lactate synthesis and pentose cycle activity [16]. However, the regulation of CDK6 in cancer metabolic reprogramming remains elusive.
Here, our data indicate that YIYA expression is correlated with recurrence in breast cancer patients. YIYA upregulation promotes tumor growth and glycolysis. Mechanistically, YIYA associates with cytosolic CDK6 to facilitate the enzymatic activation of CDK6. The presence of YIYA enhanced CDK6-dependent phosphorylation of PFKFB3, which facilitated the catalysis of F-2,6-BP. Thus, YIYA and CDK6 are shown to be required for glycolysis augmentation in breast cancer cells. YIYA knockout led to accumulation of G6P and depletion of F-2,6-BP/ F-1,6-BP in cells, as well as decreased cell proliferation, invasion and tumor growth. Our data demonstrated that the dysregulation of lncRNAs in human cancer modulates glycolysis, signifying the importance of targeting lncRNAs as an anti-glycolytic therapy.
MATERIALS AND METHODS
Clinical samples
Two sets of fresh-frozen breast cancer tissues (Yixin-Bre-01 and Duke-Bre-01) were collected from Yixing People’s Hospital (Yixing, Jiangsu Province, China) and Duke University, respectively. The study protocol was approved by the Institutional Review Board of Nanjing Medical University (Nanjing, China) and Duke University Health System. All tissue samples were collected with written informed consents. This study was carried out in accordance with the Declaration of Helsinki of the World Medical Association.
Cell culture, transfection, and lentiviral transduction.
MDA-MB-231, MCF7, BT474 and HEK293T cells, obtained from American Type Culture Collection (ATCC) in 2013, were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM)/F12 (Life technologies) with 10% Fetal Bovine Serum (FBS), and 1% penicillin/streptomycin (pen/strep). Routine Mycoplasma testing was performed by PCR. All of the cell lines were free of mycoplasma contamination tested by vendors using MycoAlert kit from Lonza tested in 2016. No cell lines used in this study are found in the database of commonly misidentified cell lines (ICLAC and NCBI Biosample) based on short tandem repeats (STR) profiling performed by vendors. Cells with relative low passage numbers (< 15) were used in the study. Plasmid transfections were performed using Lipofectamine3000 (Invitrogen). Lentiviruses were produced in HEK293T cells with ViraPower™ Lentiviral Expression System (Life Technologies) and used to transduce target cells.
RNAscope assay, immunohistochemistry and image quantification.
Detection of YIYA expression using RNAScope® probe (designed by Advanced Cell Diagnostics) was performed on breast or lung cancer cell lines or microarray of multiple cancer type tissues with RNAScope® 2.0 High Definition Assay kit according to the manufacturer’s instructions (Advanced Cell Diagnostics). Immunohistochemistry was performed as previously described [17]. The images were visualized with Zeiss Axioskop2 plus Microscope, and the slides were scanned on the Automated Cellular Image System III (ACIS III, Dako, Denmark) for quantification by digital image analysis as previous described [18]. For survival analysis, the expression of YIYA was treated as a binary variant and divided into ‘high’ and ‘low’ level. Kaplan-Meier survival curves were compared using the log rank test.
In vitro RNA-protein binding assay, RNA pull-down and mass spectrometry analysis.
In vitro RNA-protein binding and RNA pulldown assays were performed as described previously [17]. Briefly, in vitro transcribed biotinylated YIYA sense or antisense transcript were first captured by the magnetic Monoavidin beads (Bioclone Inc.) and the YIYA-bound beads were further incubated with MDA-MB-231 cell lysates. The YIYA-bound proteins/complexes were in solution digested by Immobilized Trypsin (Promega) and subjected to LC-MS/MS analysis at MD Anderson Cancer Center Proteomics Facility. For identification of binding proteins of FBXW7 and CDK6, FLAG-tagged FBXW7 and CDK6 were subjected to affinity pulldown using anti-FLAG M2 Magnetic Beads (Sigmaaldrich) using FLAG-FBXW7 and FLAG-CDK6 expressing MDA-MB-231 cells. The protein targets that associated with both FBXW7 and CDK6 were subjected to the following studies.
Cell lysis, immunoprecipitation, immunoblotting and RIP assay.
Immunoprecipitation and immunoblotting assays were performed as described previously [17]. RIP assay was performed in native conditions as previously described [17].
In vitro kinase assay.
Recombinant proteins were incubated with CDK6 + CCND3 in vitro in kinase assay buffer I (SignalChem) containing 100 μM ATP-gamma-S (ab138911) according to manufacturer’s instruction (Promega). Alternatively, the reaction was stopped, separated by SDS-PAGE and detected by Coomassie Blue staining or immunoblotting with phospho-specific antibodies.
3D spheroid proliferation assay and 3D spheroid BME cell invasion assay.
Spheroid growth of MDA-MB-231 parental or YIYA KO cells were conducted using Cultrex® 3-D Spheroid Fluorometric Proliferation/Viability Assay kit (Trevigen). 3D invasion of indicated cells were determined using Cultrex® 3D Spheroid BME Cell Invasion Assay kit (Trevigen). The changes in the area of the spheroid expansion or invasive structures were measured using ImageJ software to determine the extent of 3-D culture BME cell invasion for each sample.
Metabolic profiling and metabolite analysis of spent media.
Control, YIYA KO cells, or cells harboring CDK6 shRNA or PFKFB3 expression vectors in tetraplicate were subjected to glucose-free DMEM supplemented with 10% dialyzed serum for 16 hrs. The cells were fed back with media containing glucose or [U13C] glucose (11 mM) for 4 hrs. The metabolites were harvested and determined as previously described [19]. For metabolite analysis of spent medium, cells were seeded in 12-well plates in triplicate for 24 hr. Glucose and lactate concentrations were measured in fresh and spent medium using a Yellow Springs Instruments (YSI) 7100. The concentration of glucose and lactate were normalized to cell numbers.
CRISPR/Cas9-mediated gene editing.
YIYA knockout cell lines were generated using the CRISPR/Cas9 genome editing system. Briefly, YIYA-specific guide RNA (gRNA) expression vectors were generated as described (Sanjana et al., 2014). LentiCas9-Blast (plasmid 52962; Addgene) and gRNA cloning vector lentiGuide-Puro (plasmid 52963; Addgene) were obtained from Addgene. MBA-MD-231 cells were co-transfected with lentiCas9-Blast, gRNA1, and gRNA2 expression vectors. The transfected cells were selected using puromycin (0.5 μg/ml) and blasticidin (1 μg/ml). Isolated single colonies were subjected to detection of genomic deletions by PCR. Sequencing validation of the genomic deletions was performed after cloning the corresponding amplicon into pGEM-T-Easy (Promega).
In vivo tumorigenesis study.
All animal experiments were performed in accordance with protocol approved by the Institutional Animal Care and Use Committee of MD Anderson. MDA-MB-231 parental or YIYA KO cells (5 × 106) mixed with Matrigel at a 1:1 ratio, were orthotopically injected to the mammary fat pad of 6-week old female athymic Nu/Nu. 5 mice for each group based on power calculations, which will allow us to detect a ~30% difference in tumor growth and/or glucose uptake between groups at the 95% confidence level. Tumor size was measured weekly using a caliper. Tumor volume was calculated using the standard formula: 0.54×L×W2, where L is the longest diameter, and W is the shortest diameter.
Data analysis and statistics.
Analyses of relative gene expression were determined using the 2-ΔΔCt method with GAPDH or B2M as the internal reference genes. Results are reported as mean ± standard error of the mean (SEM) of at least three independent experiments. Comparisons were performed using two-tailed paired Student’s t test. *p < 0.05, **p < 0.01, and ***p < 0.001.
RESULTS
YIYA is required for cell proliferation and tumor growth
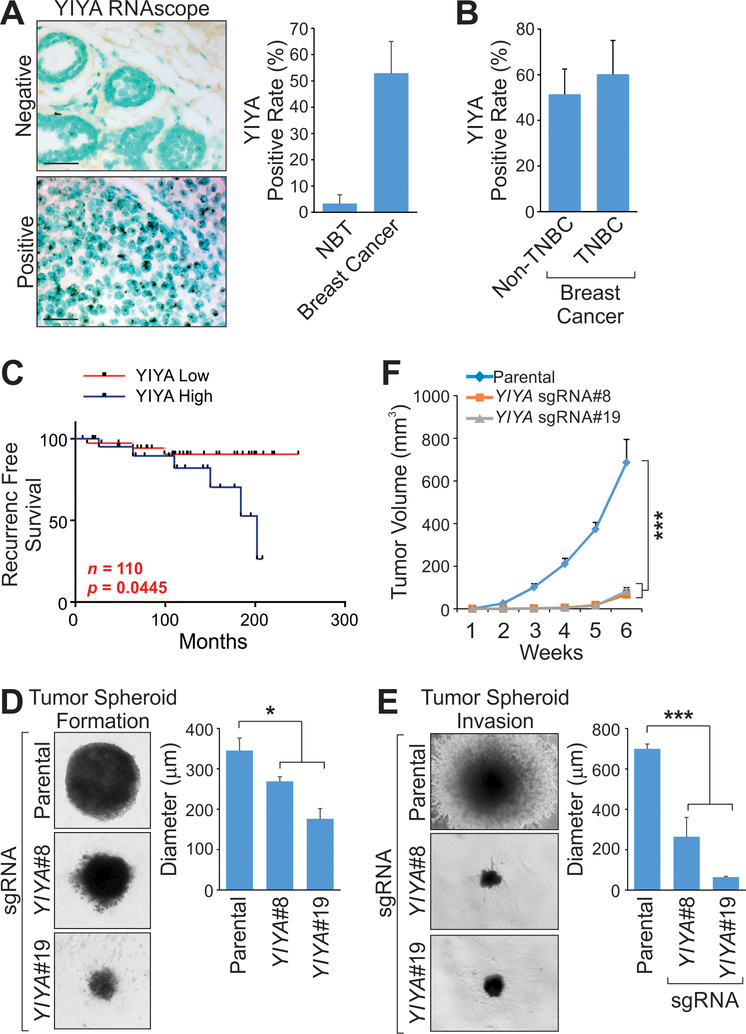
To demonstrate the functional importance of lncRNAs in breast cancer, we first determined the expression status of YIYA in breast cancer tissues with paired adjacent normal tissues using RNAscope technology, finding that over 50% of breast cancer patients expressed YIYA and less than 5% of adjacent normal tissues exhibited detectable YIYA expression (Fig. 1A and B). Breast cancer patients with high YIYA expression exhibited poor recurrence free survival (Fig. 1C). YIYA is upregulated in breast cancer cells compared to normal mammary gland epithelial cells (Supplementary Fig. S1A). Expression of YIYA regulated the cell proliferation rate, but showed minimal effects on the cell cycle (Supplementary Fig. S1B-G), suggesting that YIYA may play additional roles in promoting breast cancer cell growth that are independent to cell cycle regulation.
Figure 1.
The oncogenic role of YIYA in breast cancer. A and B, RNAscope staining of YIYA in breast tumor or adjacent normal tissues (n=35 and 130 respectively) (A) or non-TNBC and TNBC tissues (n=15 and 115 respectively) (B). Scale bar 200 μm (200X). C, Recurrence Free Survival analysis of YIYA status in breast cancer patients. (n=110, Log rank test). D and E, Tumor spheroid formation assay (D) or tumor spheroid invasion assay (E) of MDA-MB-231 cells harboring indicated sgRNAs. F, Xenograft tumor growth of mice harboring MDA-MB-231 with or without YIYA KO (n=5 animals). Error bar, SEM, n=three independent experiments (*, p<0.05, **, p<0.01, student t-test).
We generated knock out (KO) cell lines derived from MDA-MB-231 cells using CRISPR/Cas9 technology [20], using two guide RNAs (sgRNAs) flanked the transcription body of YIYA (Supplementary Fig. S1H-K). YIYA KO significantly reduced the growth and invasion of 3-dimensional tumor spheroids (Fig. 1D and E). Orthotopic injection of parental or YIYA KO cells into the mammary fat pad of mice indicated that the growth of breast tumors was significantly inhibited when YIYA was depleted (Fig. 1F). Therefore, our data demonstrates the potential oncogenic role of YIYA in breast cancer cells.
YIYA associates with CDK6 and SCF complex
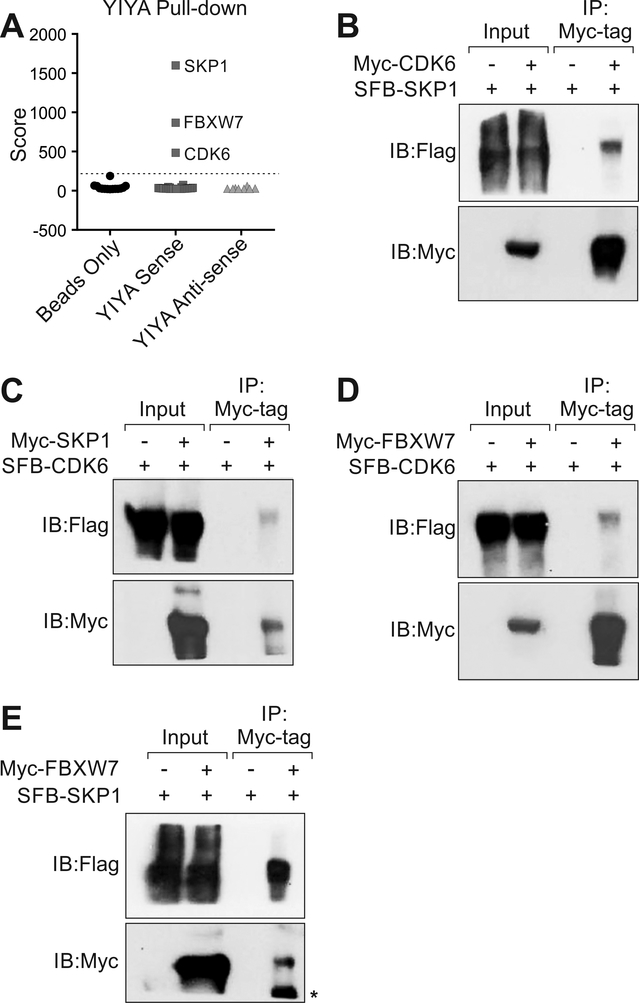
To understand the molecular mechanism by which YIYA promotes breast cancer progression, we identified the binding proteins of YIYA using liquid chromatography–mass spectrometry (LC-MS). Compared to Beads only and YIYA antisense (ant.), YIYA sense (sen.) associated with S-phase kinase-associated protein 1 (SKP1), F-box And WD repeat domain containing 7 (FBXW7), and cell division protein kinase 6 (CDK6) with high confident (Fig. 2A, Supplementary Fig. S2A). CDK6 is regulated by cyclin-D3 [21]. SKP1 and FBXW7 are both key components of Skp, Cullin, F-box containing complex (or SCF complex) [22]. We confirmed the interactions between CDK6/cyclin-D3, CDK6/SKP1 and CDK6/FBXW7 (Fig. 2B-C and Supplementary Fig. S2B). Co-immunoprecipitation also indicated that FBXW7 associated with CDK6 and SKP1 respectively (Fig. 2D and E). We then demonstrated that YIYA, CDK6 and cyclin-D3 localized to both the nucleus and the cytosol, but mostly to the cytosol (Supplementary Fig. 2C and D).
Figure 2.
YIYA mediates the interaction between CDK6 and FBXW7 complexes. A, Protein score identification in Beads only, YIYA sense (sen.) and YIYA anti-sense (ant.) samples by LC-MS. Dash line: score = 200. B-E, Immunoprecipitation followed by immunoblotting detection using indicated antibodies in MDA-MB-231 cells expressing indicated constructs.
YIYA directly associates with CDK6 and FBXW7
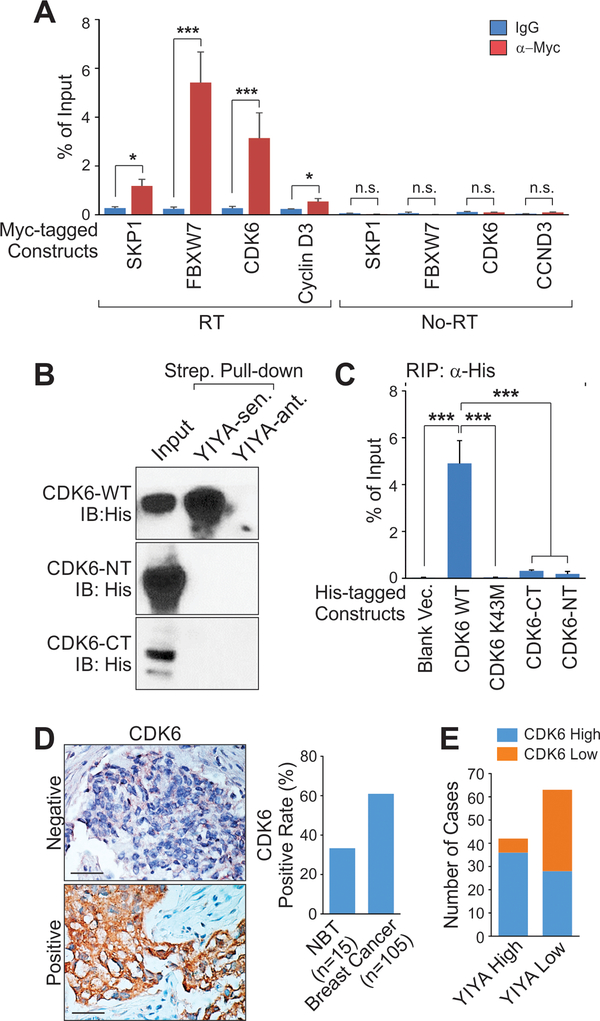
We confirmed the association between YIYA and YIYA-binding proteins in cells using an RNA immunoprecipitation assay (RIP assay) (Fig. 3A). Recombinant full-length CDK6 directly associates with YIYA sense but not the anti-sense transcript (Fig. 3B). Furthermore, neither the N-terminus, the C-terminus, nor kinase dead mutant (K43M) [23] of CDK6 exhibited interactions with the YIYA (Fig. 3B and C). The FBXW7 protein contains a WD40 domain, which has been demonstrated to be an atypical RNA-binding domain [17]. FBXW7 associated with YIYA in vitro in a WD40 domain-dependent manner (Supplementary Fig. S3).
Figure 3.
Characterization of YIYA-CDK6 and YIYA-FBXW7 interactions. A, RNA immunoprecipitation (RIP) assay followed by PCR with RT or without RT using anti-Myc antibody in MDA-MB-231 cells expressing indicated expression vectors. B, Streptavidin (Strep.) pull-down followed by immunoblotting detection using recombinant CDK6 wild type (WT), n-terminus (NT), or c-terminus (CT) in the presence of biotinylated YIYA sense or anti-sense. Bottom panel: graphic illustration of domain structure of CDK6. C, RIP assay followed by RT-qPCR detection of YIYA using anti-His antibody in in MDA-MB-231 cells expressing indicated expression vectors. D, Immunohistochemistry staining of CDK6 in breast cancer and adjacent normal tissues. Left panel: representative images. Scale Bar: 200 μm. Right panel: positive rate of CDK6 are shown, n=15, and 105 respectively. E, High or low expression of YIYA and CDK6 were plotted for correlation in breast cancer tissues. Error bar, SEM, n=three independent experiments (*, p<0.05, **, p<0.01, ***, p<0.001, student t-test).
Immunohistochemistry staining indicated that CDK6 protein is elevated in breast cancer tissues compared to normal adjacent tissues (Fig. 3D). Further, in YIYA-high breast cancer tissues, 36 over 42 cases were CDK6 high; and 35 over 63 YIYA-low breast cancer tissues were CDK6-low, suggesting the correlation between the expression of CDK6 and YIYA in breast cancer tissues (Fig. 3E).
CDK6 and FBXW7 co-regulate PFKFB3 and STK38
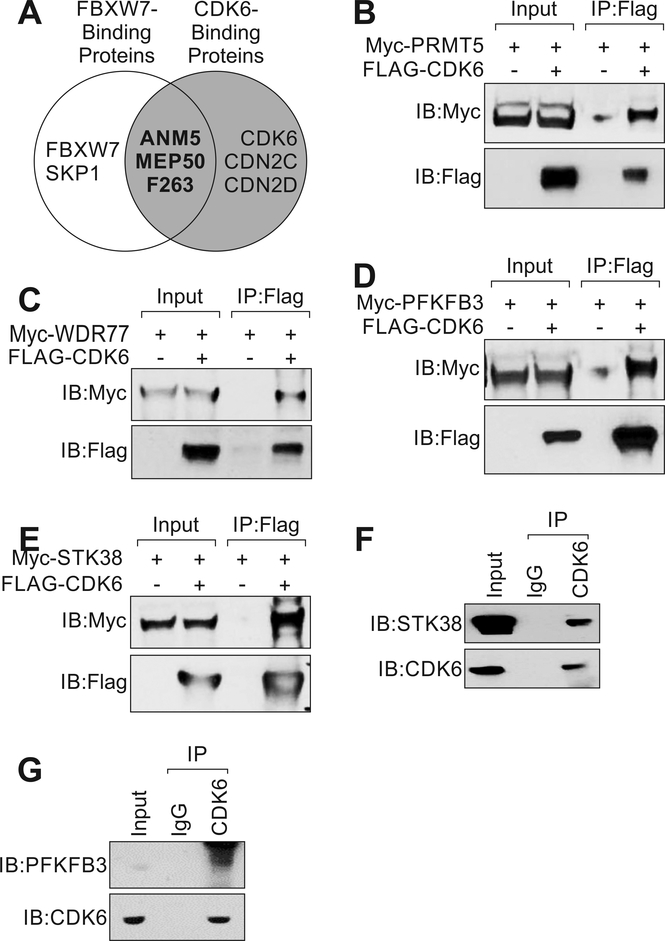
To understand how YIYA associated CDK6/cyclin-D3 and SKP1/FBXW7 complexes regulate cell growth, we identified the proteins that associated with both CDK6 and FBXW7 using FLAG-tag pulldown followed by LC-MS (Fig. 4A). The data suggested that protein arginine methyltransferase 5 (ANM5, also known as PRMT5), methylosome protein 50 (MEP50, also known as WDR77), serine/threonine kinase 38 (STK38) and 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase 3 (F263, PFK2 or PFKFB3) commonly associated with both CDK6 and FBXW7 (Fig. 4A).
Figure 4.
CDK6 and FBXW7 co-regulate PFKFB3 and STK38. A, Summery of top binding proteins associate with FBXW7 or CDK6. The proteins associates with both FBXW7 and CDK6 were shown as bold. B-E, Immunoprecipitation followed by immunoblotting detection using indicated antibodies in MDA-MB-231 cells expressing FLAG-tagged CDK6 and indicated constructs. F and G, Immunoprecipitation followed by immunoblotting detection using indicated antibodies in cell lysates extracted from MDA-MB-231.
Next, we confirmed the interactions between CDK6, FBXW7 with PRMT5, WDR77, PFKFB3 and STK38 respectively (Fig. 4B-E and Supplementary Fig. S4A-D). We also detected endogenous CDK6-STK38 and CDK6-PFKFB3 interaction (Fig. 4F and G). FBXW7 is a substrate recognition component of an SCF complex, which mediates the ubiquitination and subsequent proteasomal degradation of target proteins [24]. Overexpression of FBXW7 wild type (WT) reduced c-Myc protein levels [25, 26], which were blocked by F-box domain deletion mutant (ΔF mutant) [24], but not PFKFB3 or STK38 (Supplementary Fig. S5).
YIYA modulates CDK6-dependent phosphorylation
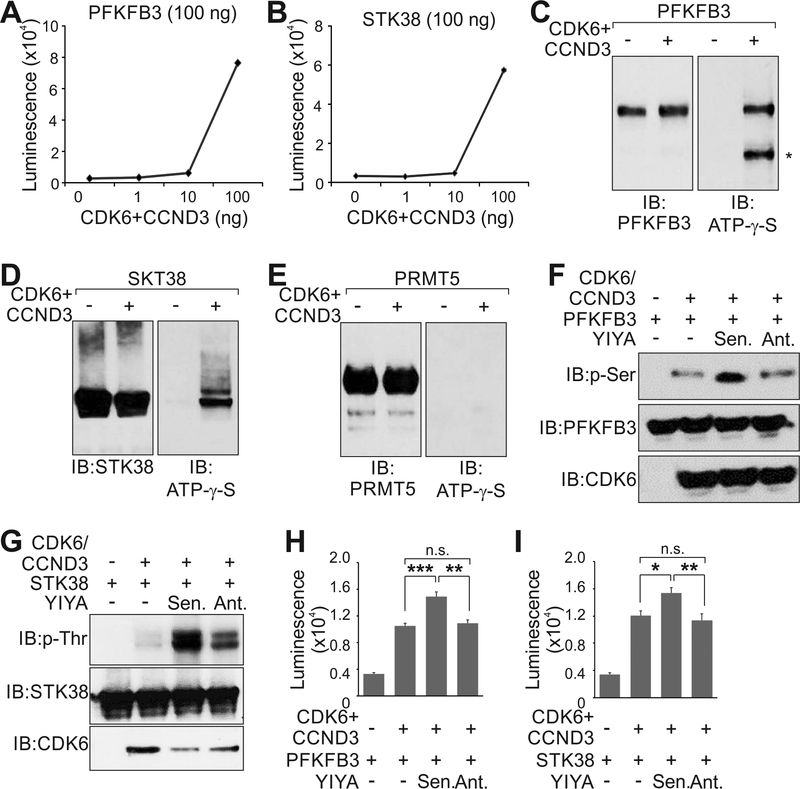
We hypothesized that the CDK6-STK38 and CDK6-PFKFB3 interactions may lead to phosphorylation of STK38 and PFKFB3. Using a quantitative in vitro kinase assay, the recombinant CDK6/CyclinD3 complex catalyzed the incorporation of ATP to recombinant PFKFB3 and STK38 in a dose-dependent manner (Fig. 5A and B), which were confirmed adenosine 5′-[γ-thio] triphosphate (ATP-γ-s) (Fig. 5C-E).
Figure 5.
CDK6 phosphorylates STK38 and PFKFB3. A and B, Phosphorylation incorporation of PFKFB3 (A) or STK38 (B) in the presence of recombinant CDK6 and CyclinD3 in dose-dependent manner. C-E, In vitro kinase assay using recombinant CDK6/CyclinD3 and indicated substrates in the presence of ATP-γ-S, followed by immunoblotting using indicated antibodies. F and G, In vitro kinase assay using recombinant CDK6/CyclinD3, PFKFB3 (F) or STK38 (G), in the presence of in vitro transcribed YIYA (sense or anti-sense), followed by immunoblotting using indicated antibodies. H and I, Phosphorylation incorporation of PFKFB3 (H) or STK38 (I) using recombinant CDK6/CyclinD3, in the presence of in vitro transcribed YIYA sense (Sen.) or anti-sense (Ant.). Error bar, SEM, n=three independent experiments (*, p<0.05, **, p<0.01, ***, p<0.001, student t-test).
LncRNAs have been reported to associate with serine/threonine protein kinases and potentially modulate their enzymatic activities [27]. The presence of YIYA sense transcript, but not the anti-sense transcript, enhanced the CDK6-dependent phosphorylation of PFKFB3 and STK38 (Fig. 5F and G). Using a quantitative in vitro kinase assay, the presence of YIYA sense significantly enhanced the CDK6-mediated phosphorylation of PFKFB3 and STK38 (Fig. 5H and I). Therefore, our data suggest that the elevated YIYA expression levels may promote CDK6 kinase activity and activate downstream signaling pathways in breast cancer cells. The enzymatic activity of CDK6 requires the presence of cyclins [21]. It is possible that the presence of YIYA promotes the association between CDK6 and cyclin D3, leading to enhanced phosphorylation of PFKFB3 and STK38.
The phosphorylation of PFKFB3 promotes glycolysis
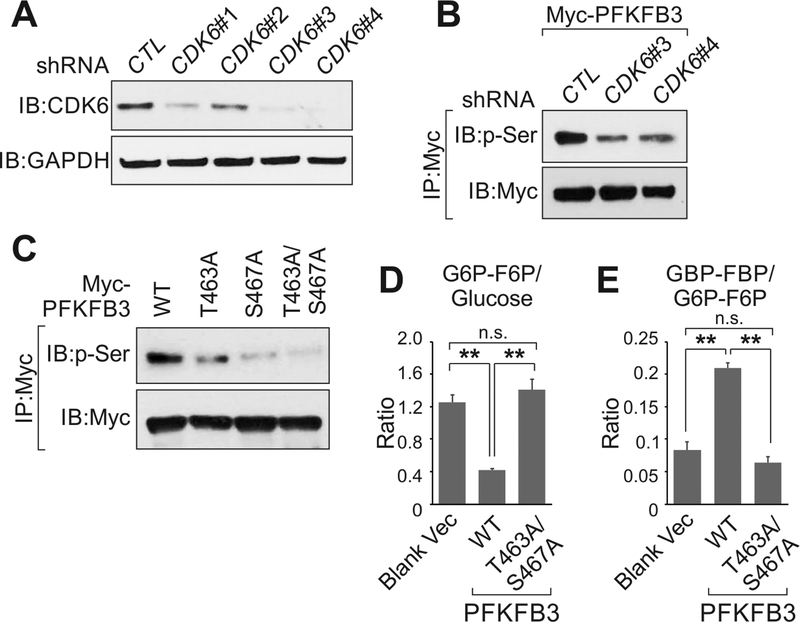
PFKFB3 catalyzes F-2,6BP [28, 29]; the latter allosterically modulates the enzymatic activity of PFK1, leading to enhanced glycolysis [30]. The enzymatic activity of PFKFB3 has been reported to be phosphorylated at residue S461, which regulates glycolysis [31]. Two phosphorylation sites (SVT463PLAS467PEPTKK), alone or in combination, were identified by mass spectrometry. To confirm the CDK6-dependent phosphorylation of PFKFB3, we generated stable cell lines with CDK6 knocked down by shRNAs (Fig. 6A). The phosphorylation of Myc-tagged PFKFB3 was reduced upon CDK6 knockdown (Fig. 6B). Expression of Myc-tagged PFKFB3 wild type, T463A, S467A or T463A/S467A mutants in MDA-MB-231 cells indicated that the phosphorylation status of exogenous PFKFB3 was impaired when T463 and S467 were both mutated (Fig. 6C). Expression of wild type PFKFB3, but not T463A/S467A mutant, enhanced the production of FBP/GBP and reduced the cellular pool of G6P/F6P (Fig. 6D and E). These data suggested that CDK6 may regulate glucose metabolism through PFKFB3 phosphorylation.
Figure 6.
PFKFB3 is regulated by CDK6-dependent phosphorylation. A, Immunoblotting detection using indicated antibodies in MDA-MB231 cells harboring indicated shRNAs. B and C, Immunoprecipitation followed by immunoblotting detection of MDA-MB-231 cells harboring indicated shRNAs (B) or expression vectors (C). D and E, Ratio of G6P-F6P over glucose (D) or GBP-FBP over G6P-F6P (E) in MDA-MD-231 expressing blank, PFKFB3 WT or T463A/S467A mutant. Error bar, SEM, n=three independent experiments (*, p<0.05, **, p<0.01, ***, p<0.001, student t-test).
In breast cancer tissues, the expression of PFKFB3 and CDK6 were both elevated in TNBC compared to other biomarker status (Gluck Breast, Oncomine) (Supplementary Fig. S6A-B). The expression of PFKFB3 significantly correlated with the expression of CDK6 in breast cancer tissues (Gluck Breast, Oncomine) (Supplementary Fig. S6C). Hence, our data suggested the molecular linkage between YIYA, CDK6 and PFKFB3 in breast cancer.
YIYA and CDK6 co-regulate glucose metabolism
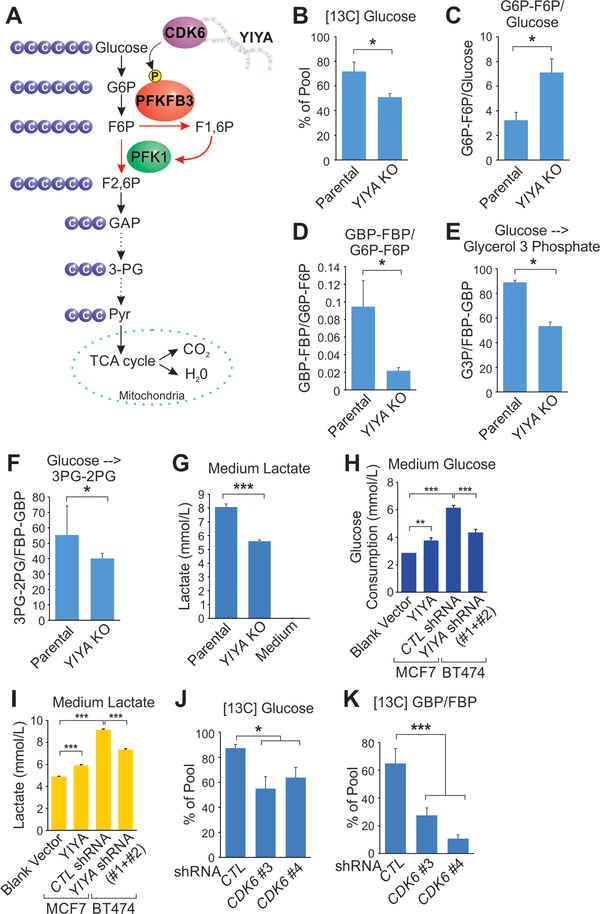
The YIYA/CDK6-dependent phosphorylation of PFKFB3 may play critical roles in regulating glucose metabolism. Therefore, we determined the metabolic flux using a 13C-based strategy (Fig. 7A). The parental and YIYA KO cells were subjected to glucose starvation followed by feedback of [U-13C] glucose. Cellular 13C-glucose uptake was impaired in YIYA KO cells compared to parental cells (Fig. 7B). Interestingly, in YIYA KO cells, we observed the accumulation of G6P/F6P and depletion of FBP/GBP, suggesting the inhibition of the conversion from F6P to FBP (Fig. 7C and D). Consistently, depletion of YIYA led to impaired production of glycerol 3 phosphate and pyruvic acid (Fig. 7E and F). The production of lactate was significantly reduced when YIYA was genetically deleted (Fig. 7G). Overexpression of YIYA in YIYA-low expressing MCF7 cells significantly enhanced glucose consumption and lactate production (Fig. 7H, I and Supplementary Fig. S7A). In YIYA-high expressing BT474 cells, YIYA knockdown repressed glucose consumption and lactate production (Fig. 7H, I and Supplementary Fig. S7B). Furthermore, knockdown of CDK6 led to decreased glucose uptake and production of GBP/FBP (Fig. 7J and K). Taken together, our data indicated that lncRNAs may be functionally important in regulating cancer metabolic reprogramming and tumor growth, making these molecules attractive therapeutic targets.
Figure 7.
YIYA/CDK6 co-regulate glucose metabolic reprogramming. A, Graphic illustration of glucose metabolic pathway regulated by YIYA and CDK6. B, The percentage of [U-13C] glucose over unlabeled glucose in MDA-MB-231 cells with or without YIYA KO were glucose starved overnight, followed with [U-13C] glucose feedback (11 mM, 4 hr). C-F, Ratio of indicated metabolites in MDA-MB-231 cells with or without YIYA KO were glucose starved overnight, followed with [U-13C] glucose feedback (11 mM, 2 hr). G, Medium lactate production detection of MDA-MB-231 cells with or without YIYA KO. H and I, Medium glucose consumption (H) and lactate production (I) of MCF7 cells expressing indicated constructs (left) or BT474 cells harboring indicated shRNAs (right). J and K, The percentage of [U-13C] glucose over unlabeled glucose (J) or [U-13C] GBP/FBP over unlabeled GBP/FBP (% of pool) (K) in MDA-MB-231 cells harboring indicated shRNAs were glucose starved overnight, followed with [U-13C] glucose feedback (11 mM, 4 hr).
DISCUSSION
The advance of next-generation sequencing has revealed large-scale dysregulation of lncRNAs in human diseases, particularly in human cancer. However, the molecular mechanisms underlying these RNA transcripts and their consequences remain limited. Our data indicates that YIYA in breast cancer may promote cancer glycolysis augmentation. Mechanistically, YIYA modulates the association between the CDK6 complex and the SCF complex in the cytosol and regulates the CDK6-dependent phosphorylation events. High YIYA expression levels promote CDK6-dependent PFKFB3 phosphorylation, leading to the enhanced phosphorylation of F6P to FBP. Consequently, expression of YIYA and CDK6 are required to maintain glycolysis at an elevated state in breast cancer. Our data demonstrate the functional role of lncRNA in breast cancer metabolic reprogramming.
The molecular mechanisms of CDK6 in cancer glycolytic reprogramming have been elusive. Based on our collected data, in the presence of YIYA, CDK6-dependent cytosolic phosphorylation events are important for promoting cancer metabolic reprogramming. The significantly elevated rates of glycolysis in breast cancer situate glycolysis as a valuable component in identifying new diagnostic strategies and therapeutic targets. Our data suggest that CDK6 inhibitors may block breast cancer glycolysis. Furthermore, anti-sense oligonucleotides targeting lncRNAs could serve as a new therapeutic strategy. Locked nucleic acids (LNA) or nanoparticle-encapsulated siRNAs targeting lncRNAs are under active development [17, 32]. Pilot data indicate that LNAs or nanoparticle-delivered siRNAs against lncRNA targets can potently inhibit tumor growth, experimental metastasis and glycolysis in vivo. Therefore, targeting lncRNAs could serve as a promising therapeutic strategy in furthering precision medicine in the near future.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr. Yin Ye and Dr. Junjie Chen for generating YIYA KO cell lines. We thank for Peter K. Park and Tina K. Nguyen for manuscript drafting. We thank the core facilities at BCM: Metabolomics Core, (NIH P30CA125123), CPRIT Proteomics and Metabolomics Core Facility (D.P.E.), (RP170005), and Dan L. Duncan Cancer Center. In addition, grants to Arun Sreekumar and Nagireddy Putluri supported this work (W81XWH-12–546 1–0130 NIH U01 CA167234 to A.S.K, R01CA220297, R01CA216426 to N.P American cancer society 127430-RSG-15–105-01-CNE to NP.). This project was also supported by the Agilent Technologies Center of Excellence in Mass Spectrometry at Baylor College of Medicine. This work was supported in part by Cancer Prevention Research Institute of Texas (CPRIT) grant number RP130397 and NIH grant number 1S10OD012304–01. This work was supported by the NCI R00 award (4R00CA166527–02), R01 award (1R01CA218036–01), CPRIT First-time Faculty Recruitment Award (R1218) and DOD Breakthrough Award (BC151465) to L.-Q.Y. and NIDDK R00 award (R00DK094981), NCI R01 award (1 R01 CA218025–01), and The CPRIT individual investigator research award (150094 and 180259) to C.-R.L.
Additional information: This work is partially supported by BCM: Metabolomics Core, (NIH P30CA125123 to A. Sreekumar), DOD W81XWH-12–1-0130 (A. Sreekumar), CPRIT Proteomics and Metabolomics Core Facility (N. Putluri) (RP170005), Dan L. Duncan Cancer Center, NIH U01 CA167234 (A. Sreekumar), ACS 127430-RSG-15–105-01-CNE (N. Putluri), R01CA220297 (N. Putluri), and R01CA216426 (N. Putluri). This project was also supported by the Agilent Technologies Center of Excellence in Mass Spectrometry at Baylor College of Medicine. This work was supported in part by CPRIT grant number RP130397 and NIH grant number 1S10OD012304–01 to D. H. Hawke. This work was supported by the NCI R00 award (4R00CA166527–02), R01 award (1R01CA218036–01), CPRIT First-time Faculty Recruitment Award (R1218) and DOD Breakthrough Award (BC151465) to L. Q. Yang and NIDDK R00 award (R00DK094981), NCI R01 award (1 R01 CA218025–01), and The CPRIT individual investigator research award (RP150094 and RP180259) to C. R. Lin. To whom correspondence should be addressed to: Chunru Lin, 1515 Holcombe Blvd, Unit 108, Room Y7.5704, Houston, TX, 77030. Email: clin2@mdanderson.org.
Footnotes
Conflict of Interest Statement: The authors declare no potential conflicts of interest.
REFERENCES
- 1.Mercer TR, Dinger ME, and Mattick JS, Long non-coding RNAs: insights into functions. Nat Rev Genet, 2009. 10(3): p. 155–9. [DOI] [PubMed] [Google Scholar]
- 2.Geisler S and Coller J, RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol, 2013. 14(11): p. 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang F and Lupski JR, Non-coding genetic variants in human disease. Hum Mol Genet, 2015. 24(R1): p. R102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fatica A and Bozzoni I, Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet, 2014. 15(1): p. 7–21. [DOI] [PubMed] [Google Scholar]
- 5.Yang F, et al. , Characterization of a carcinogenesis-associated long non-coding RNA. RNA Biol, 2012. 9(1): p. 110–6. [DOI] [PubMed] [Google Scholar]
- 6.Gatenby RA and Gillies RJ, Why do cancers have high aerobic glycolysis? Nat Rev Cancer, 2004. 4(11): p. 891–9. [DOI] [PubMed] [Google Scholar]
- 7.Marcus F, et al. , Amino acid sequence homology among fructose-1,6-bisphosphatases. Biochem Biophys Res Commun, 1986. 135(2): p. 374–81. [DOI] [PubMed] [Google Scholar]
- 8.Storey KB, Metabolic regulation in mammalian hibernation: enzyme and protein adaptations. Comp Biochem Physiol A Physiol, 1997. 118(4): p. 1115–24. [DOI] [PubMed] [Google Scholar]
- 9.Shirakihara Y and Evans PR, Crystal structure of the complex of phosphofructokinase from Escherichia coli with its reaction products. J Mol Biol, 1988. 204(4): p. 973–94. [DOI] [PubMed] [Google Scholar]
- 10.Van Schaftingen E, et al. , A kinetic study of pyrophosphate: fructose-6-phosphate phosphotransferase from potato tubers. Application to a microassay of fructose 2,6-bisphosphate. Eur J Biochem, 1982. 129(1): p. 191–5. [DOI] [PubMed] [Google Scholar]
- 11.Wu C, et al. , Roles for fructose-2,6-bisphosphate in the control of fuel metabolism: beyond its allosteric effects on glycolytic and gluconeogenic enzymes. Adv Enzyme Regul, 2006. 46: p. 72–88. [DOI] [PubMed] [Google Scholar]
- 12.Chesney J, et al. , An inducible gene product for 6-phosphofructo-2-kinase with an AU-rich instability element: role in tumor cell glycolysis and the Warburg effect. Proc Natl Acad Sci U S A, 1999. 96(6): p. 3047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clem B, et al. , Small-molecule inhibition of 6-phosphofructo-2-kinase activity suppresses glycolytic flux and tumor growth. Mol Cancer Ther, 2008. 7(1): p. 110–20. [DOI] [PubMed] [Google Scholar]
- 14.Lim S and Kaldis P, Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development, 2013. 140(15): p. 3079–93. [DOI] [PubMed] [Google Scholar]
- 15.Scheicher R, et al. , CDK6 as a key regulator of hematopoietic and leukemic stem cell activation. Blood, 2015. 125(1): p. 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zanuy M, et al. , Cyclin-dependent kinases 4 and 6 control tumor progression and direct glucose oxidation in the pentose cycle. Metabolomics, 2012. 8(3): p. 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xing Z, et al. , lncRNA Directs Cooperative Epigenetic Regulation Downstream of Chemokine Signals. Cell, 2014. 159(5): p. 1110–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li C, et al. , A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat Cell Biol, 2017. 19(2): p. 106–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ying H, et al. , Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell, 2012. 149(3): p. 656–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang H, Wang H, and Jaenisch R, Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat Protoc, 2014. 9(8): p. 1956–68. [DOI] [PubMed] [Google Scholar]
- 21.Meyerson M and Harlow E, Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol Cell Biol, 1994. 14(3): p. 2077–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cardozo T and Pagano M, The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol, 2004. 5(9): p. 739–51. [DOI] [PubMed] [Google Scholar]
- 23.Schulze-Gahmen U and Kim SH, Structural basis for CDK6 activation by a virus-encoded cyclin. Nat Struct Biol, 2002. 9(3): p. 177–81. [DOI] [PubMed] [Google Scholar]
- 24.Suryo Rahmanto A, et al. , FBW7 suppression leads to SOX9 stabilization and increased malignancy in medulloblastoma. EMBO J, 2016. 35(20): p. 2192–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moshe Y, et al. , Role of Polo-like kinase in the degradation of early mitotic inhibitor 1, a regulator of the anaphase promoting complex/cyclosome. Proc Natl Acad Sci U S A, 2004. 101(21): p. 7937–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeh E, et al. , A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol, 2004. 6(4): p. 308–18. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, et al. , LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol, 2016. 18(4): p. 431–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okar DA, et al. , PFK-2/FBPase-2: maker and breaker of the essential biofactor fructose-2,6-bisphosphate. Trends Biochem Sci, 2001. 26(1): p. 30–5. [DOI] [PubMed] [Google Scholar]
- 29.Van Schaftingen E, Fructose 2,6-bisphosphate. Adv Enzymol Relat Areas Mol Biol, 1987. 59: p. 315–95. [DOI] [PubMed] [Google Scholar]
- 30.Ros S and Schulze A, Balancing glycolytic flux: the role of 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatases in cancer metabolism. Cancer Metab, 2013. 1(1): p. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bando H, et al. , Phosphorylation of the 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatase/PFKFB3 family of glycolytic regulators in human cancer. Clin Cancer Res, 2005. 11(16): p. 5784–92. [DOI] [PubMed] [Google Scholar]
- 32.Rupaimoole R, et al. , Long Noncoding RNA Ceruloplasmin Promotes Cancer Growth by Altering Glycolysis. Cell Rep, 2015. 13(11): p. 2395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.