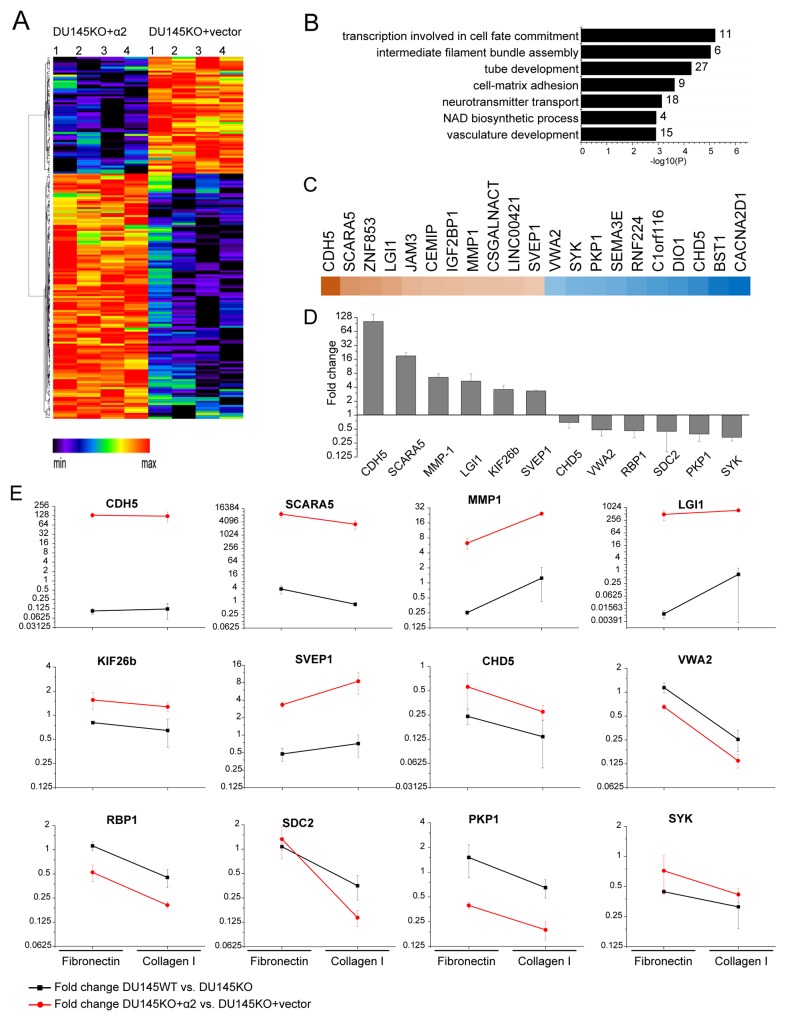
Figure 5. α2β1 integrin regulates the expression of cancer associated genes.
(A) Differential gene expression pattern of DU145KO+α2 compared to DU145KO+vector cells. Hierarchical clustering of differentially expressed (DE) genes based on relative gene expression levels detected in RNA sequencing. Red and black colors represent over and under expressed genes, respectively. (B) Differential gene expression in DU145KO+α2 compared to DU145KO+vector cells has an effect on several biological processes. Significantly over-represented gene ontology terms based on Metascape analysis. (C) Heatmap of the most up and down regulated genes in DU145KO+α2 cells when compared to DU145KO+vector cells. Brown and blue colors represent higher and lower expression levels, respectively. (D) Real time PCR analysis of selected, α2β1 integrin associated genes, in DU145KO+α2 and in DU145KO+vector cells grown as 3D spheroids. Difference is shown as the fold change of relative mRNA expression level in DU145KO+α2 compared to DU145KO+vector expression level. Data are mean (n = 3) ±SEM. (E) Effect of cell adhesion to extracellular matrix on the selected α2 integrin associated genes. Cells were grown either on collagen I or on fibronectin for 72h, after that relative mRNA levels of selected genes was analyzed by qPCR. Each data point represents the ratio of mRNA levels in α2 positive / α2 negative cells. Both, DU145KO+α2 / DU145KO+vector (red line) and DU145WT / DU145KO (black line) comparisons were made. Data are mean (n = 3) ±SEM.