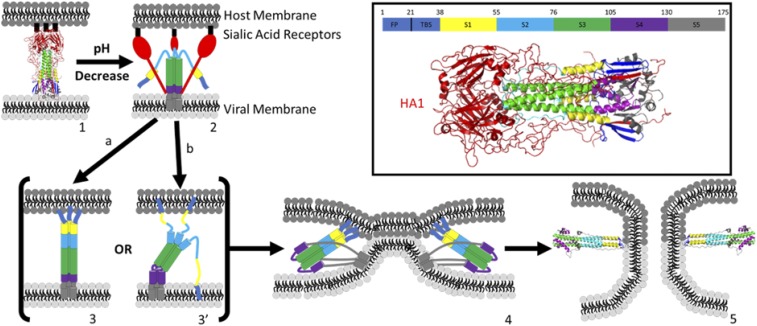
Fig. 1.
Proposed mechanism of HA-mediated membrane fusion: (1) prefusion HA structure at pH 7 (PDB ID code 1HGF); (1 → 2) low endosomal pH triggers refolding of HA, swinging the binding domain HA1 (red) away from HA2; (2 → 3) B loop refolds, extending coiled-coil structure (green), exposing the fusion peptides (FPs) (dark blue) to the host membrane; path a forms fully extended coiled coil, maintaining threefold symmetry; path b partially extends coiled coil, forming a symmetry broken intermediate [Lin et al. (13)]; (3 → 4) foldback of S4, S5 to form antiparallel coiled coil, six-helix bundle scaffold and zipping of linker leashes, bringing FP and TMD together and host and viral membranes into apposition to promote fusion; (4 → 5) pore formation, possibly facilitated by interaction between TMD and FP; structure (5) is the postfusion structure of HA (PDB ID code 1QU1). (Inset) Prefusion structure of HA (1HGF) with color legend identifying HA1 and each of the component structures of HA2, including the FP, two β-strands (TBS), helix A (S1), B loop (S2), coiled coil (S3), hinge region (S4), and ectodomain and TMD (S5).