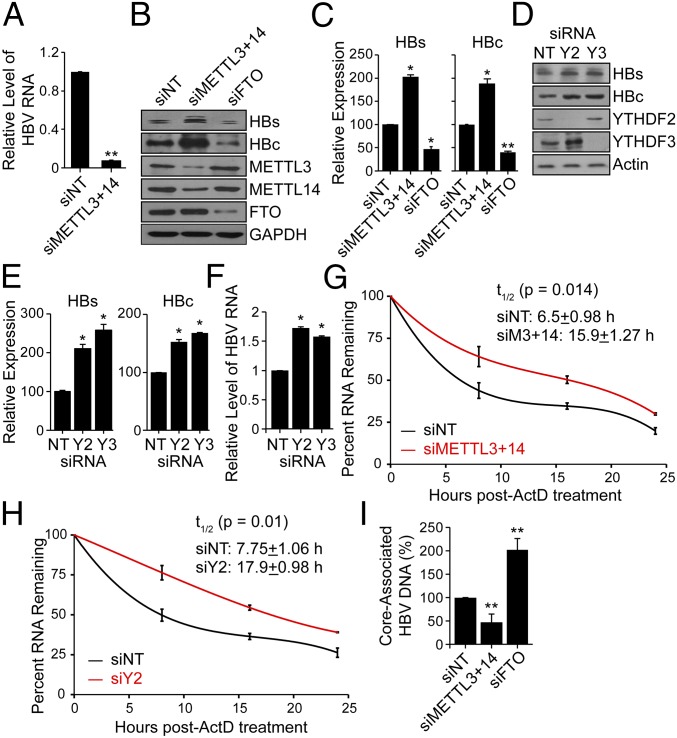
Fig. 2.
Effect of depletion of methyltransferases (METTL3/14) and demethylases (FTO) on HBV protein expression, HBV RNA stability, and reverse transcription. (A) MeRIP–RT-qPCR analysis of HBV RNA harvested from HBV-induced HepAD38 cells following siRNA depletion of METTL3 and METTL14 or nontargeting control (NT). RNA was immunoprecipitated with an anti-m6A antibody and eluted RNA was quantified as a percent of input and graphed as fraction relative to the m6A level in siNT. (B) Immunoblot analysis of HBV proteins (surface antigen, HBs) and (core, HBc) from extracts of HepG2 cells expressing the HBV 1.3mer plasmid following siRNA depletion of METTL3+METTL14, and FTO, or NT at 5 d post-HBV transfection. (C) HBV proteins levels relative to the housekeeping gene GAPDH from three independent experiments, as in B, were quantified using ImageJ. (D) Immunoblot analysis of HBs and HBc from extracts of HepG2 cells expressing the HBV 1.3mer plasmid following siRNA depletion of YTHDF2 or YTHDF3, or NT at 5 d post-HBV transfection. (E) HBV protein levels relative to the housekeeping gene Actin from three independent experiments, as in D, were quantified using ImageJ. (F) RT-qPCR analysis of HBV RNA relative to GAPDH in HepG2 cells expressing the HBV 1.3mer plasmid. At 2 d post-HBV transfection, siYTHDF2 or siYTHDF3 were added and RNA was harvested at 5 d post-HBV transfection. (G) RT-qPCR analysis of HBV RNA relative to GAPDH in the HBV 1.3mer-expressing HepG2 cells. The HBV 1.3mer-transfected HepG2 cells were depleted for METTL3 and METTL14 by siRNA, following actinomycin D treatment at 24 h post-siRNA transfection. RNA was harvested at 0, 8, 16, and 24 h post actinomycin D treatment and relative levels of remaining HBV transcript were analyzed. (H) RT-qPCR analysis of HBV RNA relative to GAPDH in the HBV 1.3mer-expressing HepG2 cells. The HBV 1.3mer-transfected HepG2 cells were depleted for YTHDF2 by specific siRNA, following actinomycin D treatment at 24 h post-siRNA transfection. RNA was harvested at 0, 8, 16, and 24 h post actinomycin D treatment and relative levels of remaining HBV transcript were analyzed. (I) Following siRNA treatment for depletion of METTL3 and METTL14, FTO, or NT in HBV-expressing HepG2 cells, the core particles were isolated (Materials and Methods). The core-associated HBV DNA was then purified and quantified by qPCR assay. The values are graphed as percent relative to the siNT, which was set at 100%. All experiments were performed in triplicate. Immunoblots shown are representative of three independent experiments, and the graph bars represent the mean ± SD of these three independent experiments. *P ≤ 0.05 and **P ≤ 0.01 by unpaired Student’s t test.