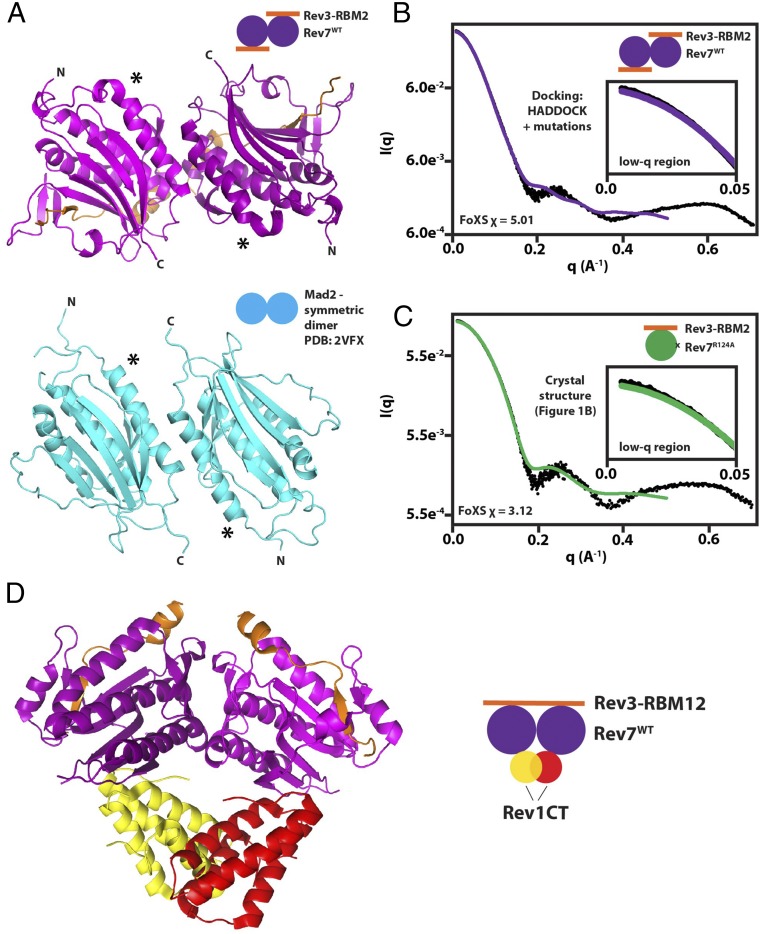
Fig. 5.
Structural modeling of the Rev7 dimer and its complex with Rev1-CT. (A) Model of Rev7WT/Rev3–RBM2 (purple) obtained using HADDOCK (57) based on mutational mapping of the Rev7 dimerization interface and its comparison with the structure of the symmetric apo-Mad2 dimer (PDB ID code 3VFX) (cyan) (48). Asterisks denote the N terminus of helix αC to demonstrate alignment. (B) Agreement between SAXS/WAXS scattering data (black dots) and scattering intensities calculated from the model of Rev7WT/Rev3–RBM2 dimer in A using FoXS (68). (C) Comparison of experimental SAXS/WAXS data with scattering data predicted from our crystal structure of the Rev7R124A/Rev3–RBM2 complex using FoXS (68). Insets in B and C show low-q regions in plots. (D) Structure of Rev7R124A (purple)/Rev3–RBM1 (orange)/Rev1-CT (yellow/red) (29) superimposed on our model of the Rev7 dimer generated using HADDOCK (57) shown in A. The Rev7 dimer is unable to bind a second Rev1-CT due to a steric clash.