Significance
Biological membranes are semipermeable barriers that are composed primarily of phospholipid bilayers or monolayers and proteins. Proteins embedded within membranes are extremely diverse in structure and function. Some membrane-integrated proteins do not fully span phospholipid bilayers but play key roles in organizing membrane surfaces. How these monolayer-integrated proteins interact with membranes has not been systematically investigated, primarily because they are very difficult to distinguish from the far more common class of proteins that fully traverse the phospholipid bilayer. We describe an approach to systematically identify monolayer-integrated proteins and demonstrate that interfacial α-helices are a structural motif that directs membrane integration for this important class of membrane protein.
Keywords: lipid droplets, membrane insertion, lipid monolayer, proteomics
Abstract
Despite not spanning phospholipid bilayers, monotopic integral proteins (MIPs) play critical roles in organizing biochemical reactions on membrane surfaces. Defining the structural basis by which these proteins are anchored to membranes has been hampered by the paucity of unambiguously identified MIPs and a lack of computational tools that accurately distinguish monolayer-integrating motifs from bilayer-spanning transmembrane domains (TMDs). We used quantitative proteomics and statistical modeling to identify 87 high-confidence candidate MIPs in lipid droplets, including 21 proteins with predicted TMDs that cannot be accommodated in these monolayer-enveloped organelles. Systematic cysteine-scanning mutagenesis showed the predicted TMD of one candidate MIP, DHRS3, to be a partially buried amphipathic α-helix in both lipid droplet monolayers and the cytoplasmic leaflet of endoplasmic reticulum membrane bilayers. Coarse-grained molecular dynamics simulations support these observations, suggesting that this helix is most stable at the solvent–membrane interface. The simulations also predicted similar interfacial amphipathic helices when applied to seven additional MIPs from our dataset. Our findings suggest that interfacial helices may be a common motif by which MIPs are integrated into membranes, and provide high-throughput methods to identify and study MIPs.
Integral membrane proteins mediate communication and solute transport between aqueous compartments and organize membrane surfaces to segregate biochemical reactions. Accordingly, proteins involved in communication or transport have one or more transmembrane domains (TMD) that physically span the phospholipid bilayer, providing a physical conduit for information or molecular translocation. In contrast, membrane-integrated proteins that organize membrane surfaces do not necessarily have to span the bilayer. Membrane-integrated proteins that lack bilayer spanning TMDs are monotopic integral proteins (MIPs). Although MIPs are found in different organellar membranes and function in diverse cellular processes, little is known about how they are targeted to, and interact with membranes.
Membrane integration occurs via direct interactions with phospholipid acyl chains, mediated either by covalent lipid modifications (1) or by regions enriched in hydrophobic amino acids, which we refer to as hydrophobic membrane domains (HMDs). HMDs spanning both leaflets of a bilayer are TMDs and have well-documented features, evidenced by over 600 solved structures in the RSCB Protein Data Bank. On the other hand, far less is known about the membrane association motifs on MIPs, which may consist of short buried loops (2–4), interfacial amphipathic α-helices (5), and hydrophobic “hairpins” (6, 7). Much of this knowledge gap stems from the difficulty in distinguishing the HMDs of MIPs from the more abundant TMDs by both biochemical and bioinformatics analyses, and the consequent dearth of high-resolution structures.
The first step in understanding the structural diversity of monotopic HMDs is to identify a sufficiently large set of proteins with unequivocal monotopic topology. In this study, we exploited the fact that lipid droplets (LDs) are enveloped by phospholipid monolayers that separate a highly hydrophobic lipid core from the aqueous cytoplasm (8). Because TMDs are flanked on both ends by soluble, hydrophilic domains, bilayer-spanning proteins are strongly disfavored from integrating into the monolayer membranes of LDs. Consequently, proteins present on the surface of LDs must have their soluble domains on the cytoplasmic side of the membrane and thus must either be peripherally attached to the membrane, via lipid anchors or interactions with another protein, or be a MIP. LDs have relatively small, well-defined proteomes (7) and because of their low densities, can be readily separated from other membrane-bound organelles by flotation on sucrose gradients (9). We took advantage of these two features to build a high-throughput assay combining quantitative proteomics and statistical modeling to comprehensively identify MIPs from LDs after removal of peripheral proteins with ionic chaotropes. Solvent-accessibility biochemical assays and coarse-grained molecular dynamics (MD) simulations on a subset of the identified MIPs revealed that these proteins interact with the membrane through an integral amphipathic helix. Combined, our results present an atlas of types of MIPs present in LDs and reveal structural details of their association with membranes, which we propose constitute a common anchoring motif for this class of proteins.
Results
Systematic Identification of LD MIPs by Quantitative Proteomics.
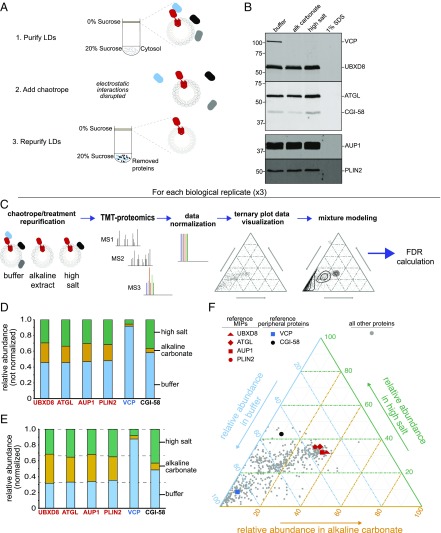
We tested the efficacy of ionic chaotropes to separate two previously well-characterized MIPs, UBXD8 (10, 11) and ATGL (12, 13), from their respective peripheral binding partners, VCP (11, 14) and CGI-58 (15) (Fig. 1 A and B). UBXD8 and ATGL were retained on LDs following treatment with ionic chaotropes but not detergent, as were two additional known MIPs, AUP1 and PLIN2. AUP1, like UBXD8, resides in both endoplasmic reticulum (ER) and LD membranes (16), and PLIN2 is thought to bind LD membranes via a hydrophobic 11-mer repeat (17, 18) directly from the cytoplasm. In contrast, the peripheral binding partners of VCP and CGI-58 were extracted by chaotropes, albeit to different extents. VCP was completely removed from LDs by both alkaline carbonate and high salt, while CGI-58 was only partially extracted by alkaline carbonate and was unaffected by high salt (Fig. 1B). This difference likely reflects the fact that VCP associates with LDs solely through electrostatic interactions with UBXD8 (11, 14, 19), whereas binding of CGI-58 to LDs requires interactions with both ATGL and membrane phospholipids (20). Although UBXD8 and ATGL were not extracted from LDs by alkaline carbonate or high salt, the absolute recovery of these two proteins and of triglycerides in the buoyant fraction was diminished after chaotrope treatment (SI Appendix, Fig. S1 A and B). Together with data from transmission electron microscopy, which revealed that LDs shrink after chaotrope treatment (SI Appendix, Fig. S1C), this suggests that peripheral proteins help maintain the size and integrity of LDs.
Fig. 1.
Chaotrope extraction and TMT proteomics used to distinguish MIPs from peripheral proteins. (A) Chaotrope treatment workflow. Purified LDs were incubated with chaotrope or detergent and repurified sucrose gradient centrifugation. Proteins extracted from the LD monolayer membrane remain in the heavy fractions while proteins anchored to the monolayer membrane float with repurified LDs. (B) MIPs are retained on LD membranes after chaotrope-treatment. Repurified LDs were analyzed by SDS/PAGE and immunoblotting with the indicated antibodies. Immunoblots from a single experiment, representative of n = 5 replicates (VCP, UBXD8, ATGL, CGI-58) or n = 2 (AUP1, PLIN2), are shown. Individual lanes were loaded based on equivalent triacylglycerol content. (C) Proteomic and data-processing workflow. Chaotrope-treated repurified LDs were subjected to TMT–based quantitative proteomics. The signals derived from the TMTs were normalized to the signals from reference MIPs characterized in B. Subsequent mixture modeling and Bayesian-based calculations were used to determine FDR values for every protein identified in all three biological replicates. (D) Nonnormalized relative abundances distinguish MIPs from peripheral proteins. Average of nonnormalized relative abundances from one biological replicate of all peptides for each indicated protein are shown. Technical duplicates were summed. (E) MIP relative abundances are at approximately equal proportions following normalization. Average of normalized relative abundances of all peptides for each indicated protein is shown. Technical duplicates were summed. (F) Ternary plot data visualization. Ternary plots have axes representing each of the three conditions. Shown is data from a single TMT experiment. Each protein is represented by a single point (gray dot) associated with three abundance values. The four validated reference MIPs have the same relative abundance in all conditions and are in the center of the plot (red symbols). VCP (blue square) is highly abundant in buffer and scarce following alkaline carbonate or high-salt extraction. CGI-58 (black dot) is abundant following buffer and high-salt extraction but at very low abundance after alkaline carbonate treatment.
To systematically identify MIPs, chaotrope-extracted LDs were subjected to quantitative proteomics using tandem mass tag (TMT) mass spectrometry (TMT-MS) (21) and the results analyzed using mixture modeling (Fig. 1C). Proteins associated with LDs after chaotrope treatment were digested with trypsin and covalently ligated with 6-plex TMT labels (21). As expected, UBXD8, ATGL, AUP1, and PLIN2 exhibited similar relative abundances after buffer and chaotrope treatments, indicating that all four MIPs remained associated with LDs to similar extents under each condition (Fig. 1D). Conversely, VCP was most abundant in buffer-treated conditions and was greatly reduced in chaotrope-treated conditions, and CGI-58 was removed from LDs by alkaline carbonate but not high salt (Fig. 1D and SI Appendix, Fig. S2A). This fully recapitulated the behavior observed by immunoblot analysis, confirming that TMT-based proteomics can be combined with chaotrope extraction to distinguish LD-associated MIPs from peripheral proteins. A complete list of all proteins identified in this study, together with their relative partitioning in buffer or chaotropes, can be found in Dataset S1.
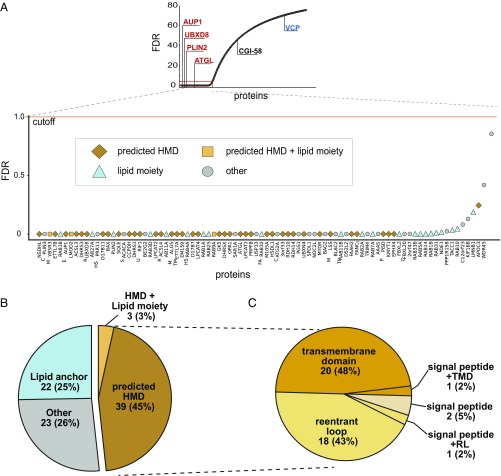
To correct for sample variability arising from variation in the efficiencies of extraction and recovery, we normalized all reporter ion signals to the average relative abundances of reference MIP peptides (Fig. 1E and SI Appendix, Fig. S2A) and used these normalized values to identify other MIPs with similar relative abundance profiles. Because, by design, the sum of all six relative abundance values must equal 1, we summed the technical duplicates to reduce the six-dimensional data to three dimensions and plotted the values on ternary plots where each axis represents the relative abundance under each extraction condition (Fig. 1F and SI Appendix, Fig. S2 B and C). Every point on the plot represents a single protein associated with three values corresponding to its relative abundance in the three experimental conditions. Proteins with equally proportioned relative abundances in the three conditions, such as the reference MIPs, cluster in the center of the plot. We fit the data to a mixture model and estimated the probability of each protein in each experiment being a member of the center cluster. We then aggregated the probabilities from the three experiments using a constrained expectation-maximization algorithm and the empirical Bayes framework to calculate a false-discovery rate (FDR) for proteins identified in all three experiments (Fig. 2A). Reference MIPs have FDR values below 2.5 × 10−5%, while the peripheral proteins VCP and CGI-58 have FDR values of 70% and 47%, respectively. These values confirm that combining chaotrope treatment, TMT-based proteomics, and our data-processing and analysis workflow, distinguishes MIPs from peripheral proteins with high confidence. Following this calibration, we used a 1% FDR cut-off to classify proteins present in our LD samples as MIPs, identifying 87 protein candidates for downstream analysis and validation (Fig. 2B and Dataset S2). The positions of the 87 candidate MIPS on the ternary plots for each TMT experiment are illustrated in SI Appendix, Fig. S3.
Fig. 2.
Bioinformatics analysis of candidate MIPs. (A) Proteins that fall below 1% FDR cut-off are candidate MIPs. The 87 proteins below 1% FDR are classified into one of four indicated groups that describe features that confer chaotrope-resistance. Classifications are based on annotations from Uniprot consortium and SPOCTOPUS (Dataset S1). (B) Pie chart representing proportions of MIPs classified as having lipid anchors, predicted HMDs, a combination of both, or neither (other). (C) Pie chart representing proportions of HMD-containing MIPs classified as TMD, reentrant loops (RL), signal peptides, or a combination of signal peptides with TMDs or RLs. SPOCTOPUS was used to distinguish between TMDs, RLs, and signal peptides.
Candidate MIPs Associate with LDs via Predicted HMDs.
Among the 87 candidate MIPs, 25 (28%) have predicted or known lipid anchors (The UniProt Consortium, 2017) (Fig. 2B and Dataset S2), and most of these are Rab GTPases that attach to membranes via lipid anchors (22). We observed that prenylation-deficient variants of GFP-Rab5 and GFP-Rab7, two hits from our high-confidence dataset, were largely absent from LD, confirming a role for lipid anchors as one mode by which MIPs are integrated into LD membranes (SI Appendix, Fig. S4 A and B). While prenylated proteins can be readily identified by the presence of a C-terminal CAAX motif (1, 23), other lipid modifications, such as palmitoylation, have no known consensus motif in eukaryotes, making it likely that that our identification of 22 lipid-anchored MIPs is an underestimate.
Next, we used SPOCTOPUS, an algorithm trained to predict TMDs and reentrant loops (24), to identify candidate MIPs that integrate into membranes via HMDs, regions enriched in hydrophobic amino acids that interact directly with lipid acyl chains. SPOCTOPUS predicted that 42 (48%) of the 87 MIPs have HMDs (Fig. 2B and Dataset S2), including the biochemically validated HMDs of UBXD8 (25) and AUP1 (16). Three MIPs—CYB5R3, ELMOD2, and Rab9—were identified in both the lipid-anchored and HMD categories, and their predicted HMDs were included in further analysis, described below. Thus, together, lipid anchors and HMDs potentially account for chaotrope resistance of 73% of our candidate MIPs. The remaining 23 proteins (26%) lacked any identifiable HMD or lipid anchor and were designated “other.”
Nearly half (19 of 42) of HMD-containing MIPs were predicted (24) to contain at least one reentrant loop, a generic term referring to V-shaped protein regions that dip into, but do not traverse, the membrane bilayer (24) (Fig. 2C and Dataset S2). Reentrant loops contribute to the formation of solvent-accessible pores in polytopic ion channels and transporters (26) but could also include the “helical hairpins” that have been proposed to embed MIPs into LD membranes (6, 7). The 19 HMDs predicted to contain reentrant loops included the experimentally validated membrane-embedded regions of UBXD8 (25) and UBXN4 (27), as well as the proposed membrane-embedded region of ATGL (13, 28). The remaining half (21 of 42) of HMD-containing MIPs were predicted by SPOCTOPUS to contain TMDs. This group includes AUP1 and ACSL3, proteins with single, experimentally validated hydrophobic domains that cannot be membrane-spanning TMDs because both termini face the cytoplasm (10, 16, 29). Of the 21 proteins with predicted TMDs, 9 have predicted TMDs on the N terminus of the protein sequence (SI Appendix, Fig. S4C), and 6 of these have been experimentally shown to be required for targeting to LDs (30–33).
Therefore, our analysis suggests that most LD-associated proteins in our dataset contain HMDs, half of which should contain bilayer-spanning domains according to state-of-the-art bioinformatics predictors. Because this membrane topology is incompatible with the phospholipid monolayers that surround LDs, we selected the prominent group of nine MIPs with N-terminal–predicted TMDs (SI Appendix, Fig. S4C) for further investigation.
The N-Terminal Region of DHRS3 Is Necessary and Sufficient for LD Integration.
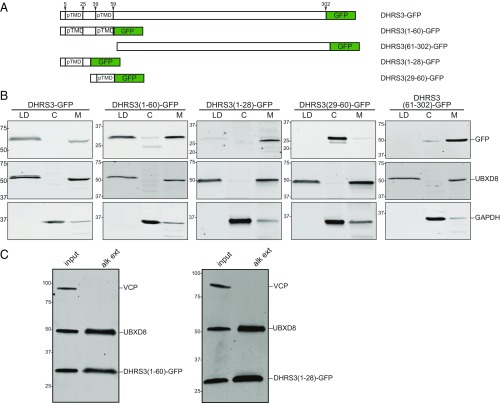
To define the function and structure of predicted TMDs on LD monolayer membranes, we assessed the topogenic features of DHRS3, a candidate MIP predicted by SPOCTOPUS to have two N-terminal TMDs (Fig. 3A). Like UBXD8 and AUP1, DHRS3 resides on both LDs and ER membranes (34) and is a lipid oxidoreductase with ∼35% sequence similarity to HSD17B11 and RDH10, also oxidoreductases and candidate MIPs containing N-terminal LD-targeting regions (30, 32) predicted by SPOCTOPUS to be TMDs. We expressed full-length DHRS3, fragments containing either (1–28, 29–60) or both (1–60) predicted TMDs individually, and the downstream sequence (61–302) as GFP fusions in HEK cells, and evaluated their steady-state localization to LDs by biochemical fractionation (Fig. 3B). GFP fusions lacking the first N-terminal predicted TMD, DHRS3(29–60) and DHRS3(61–302), did not localize to LDs and instead partitioned to the cytosol and membrane fractions, respectively. In contrast, GFP fusions to full-length DHRS3, DHRS3(1–60), and DHRS3(1–28) all localized to membrane and LD fractions. The weaker signal for DHRS3(1–28) is most likely due to the instability of this construct because the hydrophobic N-terminal region, in isolation, is likely to be a degron for the ubiquitin proteasome system (35). To verify that the N-terminal predicted TMD is integral to the LD membrane, we purified LDs from cells expressing DHRS3(1–60) and DHRS3(1–28) GFP fusions and treated them with alkaline carbonate. After repurifying the LDs, both constructs retained their interactions with the membranes, as did a positive control, UBXD8 (Fig. 3C). Taken together, these results demonstrate that the N-terminal region of DHRS3 is both necessary and sufficient for targeting and integration into LD monolayers.
Fig. 3.
N-terminal predicted TMD of DHRS3 is necessary and sufficient for targeting to LD. (A) Schematic diagram of DHRS3-GFP deletion constructs. The location of the two predicted TMDs (pTMD) is indicated. (B) Oleate-treated HEK cells expressing the indicated DHRS3 fusion constructs (A) were separated into LD, cytosol (C), and membrane (M) fractions. Representative immunoblots of these fractions probed with the indicated antibodies are shown. Blot shown is representative of at least two replicates for each construct. (C) Chaotrope-resistant integration of DHRS3(1–60) and DHRS3(1–28) fusions into LD. Purified LDs (input) were treated with alkaline carbonate and repurified on a second sucrose gradient and analyzed by immunoblotting, as in Fig. 1. Representative immunoblots (n = 3) with the indicated antibodies are shown.
The N Terminus of DHRS3 Contains a Partially Buried α-Helix.
Because TMD topology, with soluble regions on both sides of a membrane, cannot, by definition, be an option for LD-integrated proteins, the predicted TMDs in our dataset likely represent a topological motif that SPOCTOPUS is misclassifying. Alternatively, these domains can have hybrid topologies, adopting a TMD topology in a bilayer and a different topology in monolayers. To distinguish between these possibilities, we mapped the solvent accessibility of the N-terminal region of DHRS3 (residues 2–58) in ER bilayer and LD monolayer membranes by assessing the reactivity of single-cysteine mutants to methoxypolyethylene glycol maleimide (mPEG) (36). This membrane-impermeable molecule reacts covalently and exclusively with the thiol group of free cysteines.
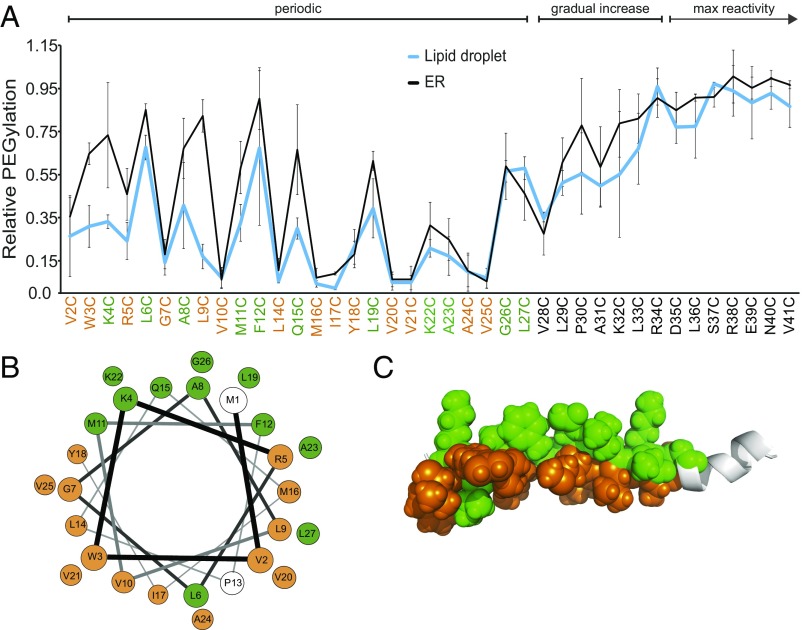
We generated a library containing DHRS3-GFP single cysteine mutants at each position within the N-terminal 60 amino acids, and reacted LD (SI Appendix, Figs. S5 and S7) and ER (SI Appendix, Figs. S6 and S7) membranes from cells expressing each construct with mPEG. The reactivity of each cysteine mutant was normalized to the reactivity in the presence of Triton X-100, a measure of the maximum reactivity of each individual mutant. Plotting the reactivity values for each mutant showed three distinct regions on the protein sequence (Fig. 4A). The first 27 residues exhibited a periodicity strikingly consistent with a partially buried α-helix (Fig. 4A). Note that residue P13 is absent from the plot because the mutated residue (P13C) did not label in the presence or absence of Triton X-100 (SI Appendix, Figs. S5 and S6). Then, residues 28–34 exhibited a gradual increase in reactivity and decreased periodicity, consistent with a helix emerging from the membrane into bulk solvent (i.e., cytosol). Finally, residues 35–58 exhibited maximum reactivity and no periodicity, consistent with complete solvent exposure (Fig. 4B and SI Appendix, Figs. S5–S7). Importantly, the pattern of reactivity was indistinguishable between purified ER and LD membranes, indicating that the DHRS3 N terminus adopts the same topology in both bilayer and monolayer membranes. Plotting the reactivity of residues 1–26 on a helical wheel (Fig. 4B) or an α-helical peptide model (Fig. 4C) revealed clear amphipathic periodicity, supporting the conclusion that DHRS3 is anchored to both the ER bilayer and the LD monolayer by a partially buried α-helix and not by a TMD, as predicted by SPOCTOPUS.
Fig. 4.
The N-terminal predicted TMD of DHRS3 forms a partially buried amphipathic α-helix. (A) Relative mPEG reactivity of systematic single cysteine-mutants of DHRS3(1–60) GFP. Each point represents mean ± SD from n = 3 independent experiments. Green and orange labels identify exposed and buried residues, respectively. “Periodic,” “gradual increase,” and “max reactivity” describe the reactivity pattern of the indicated region. (B) DHRS3(1–26) modeled onto a helical wheel and color-coded green for exposed residues and orange for buried, as in A. (C) DHRS3(1–35) modeled onto a peptide with green and orange spheres indicating exposed and buried residues, respectively. Residues 27–35 are shown in gray ribbon.
MD Simulations Predictions Support Interfacial Amphipathic α-Helices in MIPs.
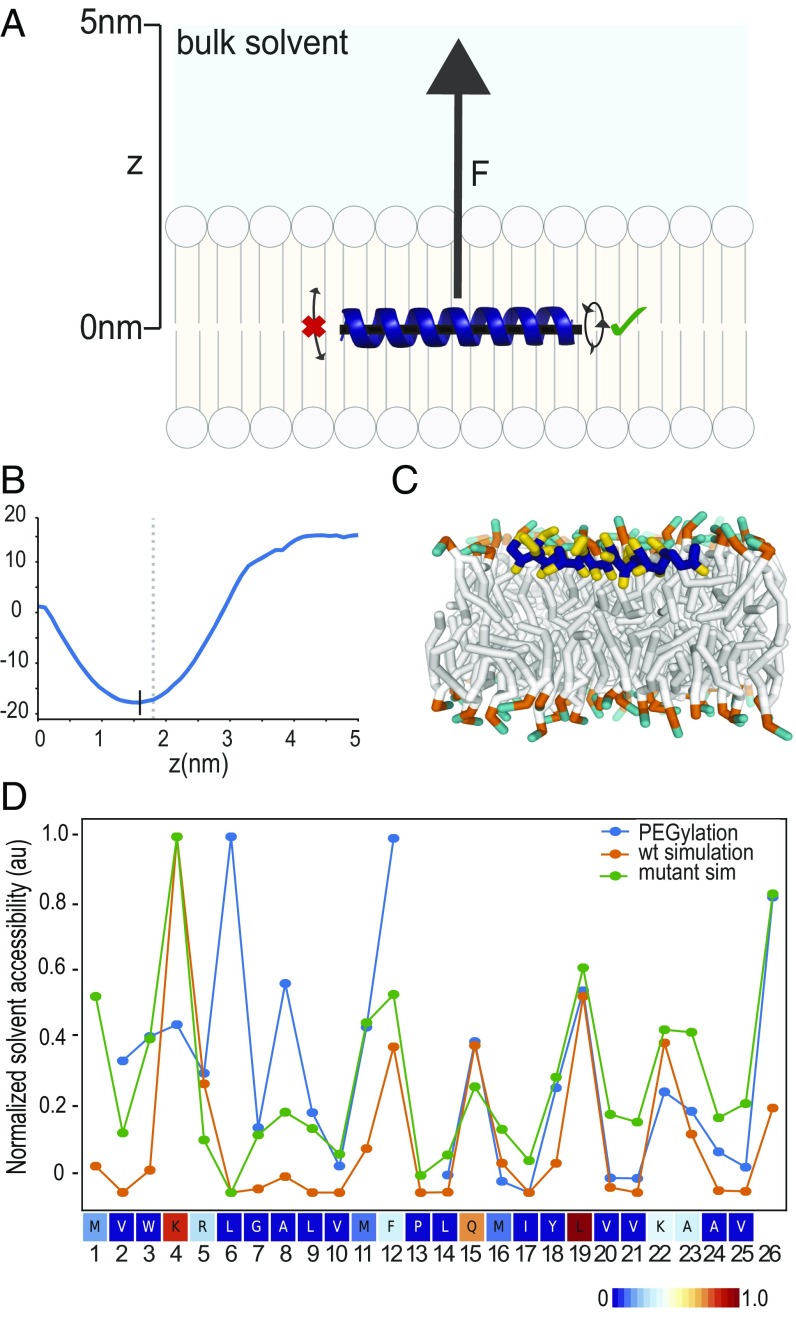
We used coarse-grained MD simulations in a phosphatidylcholine (POPC) bilayer to investigate the depth and orientation at which the N-terminal α-helix of DHRS3 is buried in the membrane. Specifically, we performed umbrella sampling simulations of a helical model of DHRS3(1–27) to calculate the change in free energy as a function of distance to the center of the lipid bilayer and generate a potential of mean force (PMF) (Fig. 5A). The free-energy minimum was found to be at ∼1.6 nm from the center of the membrane (Fig. 5B), allowing some side-chain atoms to be at the membrane–solvent interface (Fig. 5C) in agreement with the PEGylation data.
Fig. 5.
MD simulations predict DHRS3 N terminus to be at membrane-solvent interface. (A) Schematic of the steered simulation for calculating the PMF of a peptide at various depths of a bilayer membrane. An α-helical peptide was placed in the middle of a bilayer membrane and a force (F) applied in the positive (Z) direction. During the simulation, the peptide was not allowed to tilt, but was allowed to rotate. (B) Graph of the PMF measurements for DHRS3(1–26) as it is pulled from the middle of the membrane toward bulk solvent. The black vertical line denotes the minimum PMF force, and thus, the most stable location of the peptide in the membrane. The gray vertical dashed line denotes the top of the membrane. (C) Cross-sectional view of DHRS3(1–26) during an unrestrained simulation in the membrane at the location indicated by the black vertical line in B. (D) Side-chain solvent accessibility measurements from MD simulations corroborate solvent accessibility measurements from biochemical assay. DHRS3(1–26) wild-type and individual cysteine mutants were placed in the membrane position corresponding to the PMF minimum in B and allowed to run without any restraints. The calculated accessibilities of each amino acid in wild-type and individual cysteine mutants are shown in a line plot together with data from the PEGylation experiment (Fig. 4A), normalized to values between 0 and 1. Calculated accessibilities of wild-type DHRS3(1–25) is also rendered as a heatmap.
Then, to obtain a more quantitative description of the peptide topology, we calculated solvent accessibilities of each individual amino acid side-chain during an unrestrained simulation of the same helical model of DHRS3(1–27), starting from the conformation at the free-energy minimum. The average accessibilities show a periodic pattern of alternating exposed and buried residues, corresponding extremely well with the accessibility values determined from the PEGylation experimental data (Fig. 5D), except for residues Trp3, Leu6, and Ala8. To test if these discrepancies could be caused by the mutation of these amino acids to cysteine, which has a polar side-chain, we generated models of single cysteine mutants of the DHRS3(1–27) peptide in silico and repeated the same unrestrained simulation and solvent exposure calculation as for the wild-type sequence (Fig. 5D). The exposure of Trp3 and Ala8 increased when mutated to a cysteine, corroborating the PEGylation data and suggesting that when Trp3 and Ala8 are mutated to cysteines, they are more exposed. In contrast, mutating Leu6 to a cysteine did not increase exposure. The weaker correspondence between experimental and simulation accessibilities for the first residues of the peptide can be attributed to conformational dynamics of this region, as hinted by the large SDs of the PEGylation data for residues 2–5 (Fig. 4A), as well as to limitations of the coarse-grained model used in the simulations. The MARTINI model constrains backbone dihedral angles to their starting values, meaning the entire peptide is kept in a rigid α-helical conformation. Although our simulations used a reduced force constant for these constraints, to allow for some flexibility, it is unlikely they capture realistic bending of the peptides, which could explain the discrepancies between experimental and simulated accessibilities. Moreover, the biophysical properties of the extreme N terminus of DHRS3 could be disrupted by a cysteine substitution at Leu6. Because the peptide is at the interface of the membrane and the solvent, it is not unreasonable to propose the breaking of the α-helical hydrogen bonds that would otherwise come at too high a cost if buried deeper in the membrane. In support of this interpretation, a study of interfacial domains of polytopic proteins demonstrated that 70% of the structures are unstructured and only 30% α-helical (37). Taken together, the experimental and simulation results confirm that the DHRS3 N terminus is a partially buried amphipathic helix at the interface of the membrane and the cytosol.
To determine if other predicted TMDs among our candidate MIPs could also adopt similar interfacial topologies, we subjected helical models of the other selected eight N-terminal predicted TMDs predicted to be α-helical (SI Appendix, Fig. S3C) to the same simulation protocol and rendered the solvent accessibilities for each amino acid as heatmaps for direct comparison (SI Appendix, Fig. S8). Six of the proteins with predicted N-terminal TMDs showed PMF profiles and minima similar to those of DHRS3 (SI Appendix, Fig. S9). The solvent accessibility calculations for five of these predicted TMDs also exhibited alternating solvent-exposed and buried residues at the membrane surface, corroborating their similarity to DHRS3 and supporting a model in which they are embedded in the membrane by interfacial amphipathic helices (SI Appendix, Fig. S8A). Two other proteins with predicted N-terminal TMDs, HSD17B11 and RDH14, show free-energy minima at the membrane center (z = 0.0 nm), indicating a preference for a fully buried topology (SI Appendix, Fig. S9). However, during the unrestrained simulations, HSD17B11 moved closer to the surface of the bilayer and interacted with the phospholipid head groups via a few charged side-chains (SI Appendix, Fig. S8B). FAAH2 also moved similarly along the bilayer normal during the unrestrained simulation, designating FAAH2 and HSD17B11 as subsurface buried helices that still exhibit alternating solvent-exposed and buried amino acids, likely with exposed residues that interact more with the phospholipid head groups than with solvent. In contrast, RDH14 adopted a TMD topology during the unrestrained simulation, with the ends of the helix exposed to solvent on either side of the membrane (SI Appendix, Fig. S8C), consistent with its calculated free-energy minimum in the middle of the membrane. This topology is, however, unlikely to exist in both membrane bilayers and LDs, given the hydrophobic interior of the latter. To test the possibility of RDH14 adopting different topologies in lipid bilayers and droplet monolayers, we ran umbrella sampling and unrestrained simulations, as before, but using a membrane of DXPC lipids (corresponding to atomistic C24:0 dilignoceroyl–C26:0 dihexacosanoyl tails), whose tails are substantially longer than POPC (∼55–60 Å vs. ∼40 Å for POPC). As such, the RDH14 peptide (∼55 Å in length) cannot adopt a transmembrane topology and one of its termini must be exposed to the hydrophobic membrane interior. The PMF calculated from these simulations showed a minimum at 2.5 nm (SI Appendix, Fig. S12A), placing RDH14 below the phospholipid heads and parallel to the membrane plane, similar to DHRS3. During the unrestrained simulation, the peptide remained at the membrane–solvent interface, its side-chains showing an alternating pattern of solvent exposure (SI Appendix, Fig. S12B).
These findings strongly support the conclusion that that DHRS3 is anchored to membranes by an interfacial amphipathic α-helix at the solvent–membrane interface, a configuration that is likely to be adopted by five other MIPs from our study that were incorrectly predicted to contain TMDs by SPOCTOPUS. RDH14 represents a unique case of apparent dual-topology, depending on the type of membrane it is inserted in, which might explain some of the difficulties in distinguishing MIPs from regular TMDs with prediction algorithms. Given the approximate nature of the simulations and the limitation of the coarse-grained model, more work is necessary to confirm the predicted membrane topologies of HSD17B11 and FAAH2.
Discussion
Identification of MIPs by Quantitative Proteomics and Mixture Modeling.
Although MIPs are an important class of integral membrane proteins present in all cellular membranes, it is difficult to distinguish them biochemically from bilayer-spanning proteins. Consequently, fewer than 50 MIPs have been unambiguously identified (blanco.biomol.uci.edu/mpstruc/), and our current understanding of the structural mechanism by which they interact with membranes is poor. In this study, we exploited the fact that all integral proteins in LDs must be MIPs because these organelles lack phospholipid bilayers.
Approximately half of the 87 high-confidence hits identified in this study lack a predicted HMD and half of these are proteins with an identifiable lipid-anchor motif. Those proteins lacking both HMDs and lipid-anchoring motifs were designated as “other” and likely include proteins anchored via a lipid moiety that cannot be predicted with existing tools and proteins that associate with LDs through high-affinity interactions with MIPs that cannot be completely disrupted by ionic chaotropes. For example, UBE2G2, a protein assigned to this class because it lacks any identifiable HMD, is a well-characterized peripheral protein that binds with high affinity (Kd = 20 nM) (38) to AUP1, one of our “reference” MIPs. Some proteins assigned to the “other” category could associate with LDs via noncanonical HMDs that are not detected by current algorithms. For example, perilipin 3 (PLIN3), identified in this study, interacts with LDs via 11-mer repeats that are unstructured in solution but adopt an 11/3 helical conformation at the LD surface (17). Finally, the “other” category is likely to include some abundant cytosolic proteins that nonspecifically associate with LDs and are inefficiently dissociated by chaotropes.
The HMD category contains most previously identified LD proteins with well-characterized membrane-embedded domains. Of the 42 proteins with predicted HMDs, 16 have empirical evidence of being membrane-embedded (Dataset S2), and two, PKMYT1 and BAX, were recently found on LDs (39). PKMYT1, a kinase that phosphorylates Cdc2 and localizes to the ER (40), is predicted to have a reentrant loop (Dataset S2), consistent with its ability to traffic from the ER to LDs. BAX, a proapoptotic protein that permeabilizes mitochondrial outermembranes by inserting hydrophobic helices into the membrane upon oligomerization (41, 42), is one of three mitochondrial proteins (in addition to OPA1 and GK5) that we identified in this category. Of these, OPA1 is the only one with a documented function on LD membranes, where it serves as an A-kinase anchoring protein and is necessary for hormonal control of perilipin phosphorylation and lipolysis (43). It is possible that proteins like BAX and GK5 could be sequestered on LDs for “storage,” as in the case of histones H2A, H2B, and H2Av that are attached to LDs in Drosophila embryos, apparently to be used for chromatin assembly during times of high demand (44).
Amphipathic α-Helices Mediate MIP Integration into LD and ER Monolayers.
The alternating pattern of solvent accessibility of the N-terminal HMD of DHRS3, revealed by our PEGylation experiments, agrees surprisingly well with that of an amphipathic α-helix (Fig. 5). This finding was unexpected because the HMDs of proteins with dual localization to LD and the ER are proposed to adopt helical “hairpin” topologies (6). Other MIPs that have been reported to interact with the LD surface via amphipathic α-helices differ substantially from the examples identified here as they bind to LD membranes directly from the cytoplasm and are not predicted by SPOCTOPUS to be TMDs (45, 46). Unlike DHRS3, which is stably integrated, binding of CCT-α to LD membranes is reversible and dependent on macromolecular crowding on the membrane surface (45, 47). These helices can also be distinguished by their higher overall hydrophobic dipole moment, a metric of the axial distribution of hydrophobic and hydrophilic amino acids along a helix (48). The amphipathic helices on CCT-α and CIDEA have hydrophobic moments of 0.48 (49) and 0.50 (50), respectively. These values are considerably higher than DHRS3 and the N-terminally targeted MIPs identified in this study, all of which have hydrophobic moments of <0.26 (Dataset S3). The smaller hydrophobic moment of these N termini indicates a larger axial hydrophobic surface compared with CIDEA and CCT-α, suggesting more extensive interactions with phospholipid acyl chains. Moreover, the higher number of charged side-chains along the polar faces of CIDEA and CCT-α suggests a more superficial engagement of these helices with the membrane, consistent with their solubility in cytoplasm-reversible binding to LDs. Hydrophobic moments can thus be used to distinguish reversibly binding amphipathic helices from stably bound integral amphipathic helices. However, because TMDs can have similar ranges of hydrophobic moments (51), distinguishing integral amphipathic helices from TMDs will require the identification of other distinguishing features.
Interfacial amphipathic helices need not be restricted to N termini. UBXD8, for example, inserts into LD via an internal HMD that is proposed to form a hydrophobic hairpin (11, 25, 52) with a hydrophobic moment value of 0.25 (Dataset S3). We speculate that UBXD8 may also be anchored to LD membranes by an integral interfacial amphipathic helix. Further investigation will be required to determine whether this motif is more broadly applicable to MIPs with internal anchors and whether helical hairpins indeed exist in membrane monolayers.
In conclusion, the work presented here provides a high-throughput quantitative method to identify and classify membrane-integrated proteins. Our combined experimental and computational analysis of the topology of the N terminus of DHRS3 sheds light on an understudied class of membrane topologies. Finally, our dataset of high-confidence MIPs comprises a valuable resource for the development of better computational tools that predict different types of membrane topologies.
Materials and Methods
SI Appendix, Supporting Materials and Methods provides information on cell culture, transfection, drug treatment, antibodies and reagents, plasmids, electron microscopy, immunoblotting, cell fractionation, solvent accessibility assays, TMT-MS sample preparation, structural modeling and MD simulations, and statistical methods.
LD Purification and Chaotrope Treatment.
Sixteen 15-cm plates were purified as described previously (9); after 12–16 h of treatment with 200 μM Oleate, cells were incubated in hypotonic lysis buffer (HLB) [20 mM Tris⋅HCl (pH 7.4), 1 mM EDTA] for 10 min, dounce-homogenized, and centrifuged at 1,000 × g for 10 min. The postnuclear supernatant was transferred to a new tube, adjusted to a final concentration of 20% (wt/wt) sucrose, and overlaid with 3 mL of HLB containing 5% (wt/wt) sucrose and then with 3 mL of HLB. After centrifugation at 28,000 × g for 1 h using a SW41 rotor, the buoyant LD-enriched fraction was collected using a Beckman tube slicer. TAG content was assayed by a Serum Triglyceride Determination Kit (Sigma). Next, 300 μg of TAG content was brought to a final concentration of 0.5 μg/μL and then doubled in volume with a 2× solution of each chaotrope in HLB. The tubes were inverted and placed on ice for 30 min. The solution was brought to a final concentration of 20% (wt/wt) sucrose, and overlaid with 1 mL of HLB containing 5% (wt/wt) sucrose and then with 1 mL of HLB. After centrifugation at 28,000 × g for 1 h using a SW60 rotor, the buoyant LD-enriched fraction was collected using a Beckman tube slicer. Proteins were solubilized for 10 min in 2% TritonX-100 at 65 °C, precipitated with 15% TCA, and washed twice with cold acetone. Proteins were either resolubilized in laemmli buffer with β-mercaptoethanol and analyzed by SDS/PAGE/immunoblotting or resolubilized with 8 M urea, 0.25% RapiGest (Waters) in 100 mM ammonium bicarbonate (pH 8.0) for TMT-MS sample processing.
Mass Spectrometry.
Samples were analyzed by online capillary nano–LC-MS/MS. Peptides were separated on an in-house–made 20-cm reverse-phase column [100-µm inner diameter, packed with ReproSil-Pur C18-AQ 3.0 µm resin (Dr. Maisch GmbH)] equipped with a laser-pulled nanoelectrospray emitter tip. Peptides were eluted at a flow rate of 400 nL/min using a 180-min gradient (buffer A: 0.2% formic acid and 5% DMSO in water; buffer B: 0.2% formic acid and 5% DMSO in acetonitrile) in an Eksigent ekspert nanoLC-425 system (AB Sciex). Peptides were then analyzed using a LTQ Orbitrap Elite mass spectrometer (Thermo Scientific). Data acquisition was executed in data-dependent mode with full MS scans acquired in the Orbitrap mass analyzer with a resolution of 60,000 and m/z scan range of 400–1,300. The top 10 most-abundant ions with intensity threshold above 500 counts and charge states 2 and above were selected for fragmentation using collision-induced dissociation (CID) with isolation window of 1.2 m/z, collision energy of 35%, activation Q of 0.25, and activation time of 10 ms. The CID fragments were analyzed in the ion trap with rapid scan rate. Dynamic exclusion was enabled with repeat count of 1 and exclusion duration of 45 s. The AGC target was set to 1,000,000 and 5,000 for full Fourier Transform MS (FTMS) scans and ITMSn scans. The maximum injection time was set to 250 s and 100 s for full FTMS scans and ITMSn scans. The MS3 was performed in a customized “multi-notch” mode where the top five fragment ions from MS2 were selected synchronized for MS3 fragmentation by higher-energy collisional dissociation (HCD), as previously described (53).
Raw data files were converted to mzXML files using msconvert (proteowizard.sourceforge.net/tools.shtml). The resulting files were searched against a “target-decoy” sequence database (54) consisting of the Uniprot human database (downloaded June 11, 2013 and containing 88,902 entries), common contaminate proteins, and the corresponding reversed sequences using the SEQUEST algorithm [SEQUEST v.28 (rev. 12)]. The parent mass tolerance was set to ±10 ppm and the fragment mass tolerance to 0.6 Da. Enzyme specificity was set to trypsin and maximum number of missed internal cleavage was set to 3. Oxidation of methionines was set as variable modification. Carbamidomethylation of cysteines and TMT modification on the peptide N terminus and lysine were set as variable modifications. Data were then filtered to a 1% peptide using a linear discriminator analysis (55–57). Quantification of TMT reporter ions was performed as previously described (53).
MS Data Processing.
All data processing was done in Python and the script is available at https://github.com/camisap/MIPome_analysis. Briefly, all peptides identified in each proteomic experiment were verified as fully labeled by the TMT isobaric labels by a searching for modifications indicated in the peptide sequence output by Sequest. All peptides not fully labeled, or with summed signal intensities <1,000 from all six TMT labels, or with isolation specificity below 0.8, were eliminated from the dataset. All keratin peptides were also eliminated from all analysis. All signals from each TMT label were normalized to the average relative abundance of ATGL peptides corresponding to each label.
Supplementary Material
Acknowledgments
We thank James Olzmann, Kirill Bersuker, and Colin Gottlieb for critical reading of the manuscript; and Jon Mulholland for help with electron microscopy. This work was supported National Institute of General Medical Sciences Grants R01GM074874 (to R.R.K.), 5R01EB001988 (to T.H.), and R35GM122543 (to M.L.). T.H. and J.Q. were partially supported by grants from the National Science Foundation (DMS-1407548). C.I.P. was supported by a training grant from National Institute of General Medical Sciences (T32GM007276) and the Graduate Research Fellowship Program from the National Science Foundation. J.R. was supported by a Niels Stensen postdoctoral fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All data processing was done in Python and the script is available at https://github.com/camisap/MIPome_analysis.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807981115/-/DCSupplemental.
References
- 1.Ray A, Jatana N, Thukral L. Lipidated proteins: Spotlight on protein-membrane binding interfaces. Prog Biophys Mol Biol. 2017;128:74–84. doi: 10.1016/j.pbiomolbio.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Gamsjaeger R, et al. Membrane binding of beta2-glycoprotein I can be described by a two-state reaction model: An atomic force microscopy and surface plasmon resonance study. Biochem J. 2005;389:665–673. doi: 10.1042/BJ20050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Del Papa N, et al. Human beta 2-glycoprotein I binds to endothelial cells through a cluster of lysine residues that are critical for anionic phospholipid binding and offers epitopes for anti-beta 2-glycoprotein I antibodies. J Immunol. 1998;160:5572–5578. [PubMed] [Google Scholar]
- 4.Szeto TH, Rowland SL, Rothfield LI, King GF. Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc Natl Acad Sci USA. 2002;99:15693–15698. doi: 10.1073/pnas.232590599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shiba T, et al. Structure of the trypanosome cyanide-insensitive alternative oxidase. Proc Natl Acad Sci USA. 2013;110:4580–4585. doi: 10.1073/pnas.1218386110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kory N, Farese RV, Jr, Walther TC. Targeting fat: Mechanisms of protein localization to lipid droplets. Trends Cell Biol. 2016;26:535–546. doi: 10.1016/j.tcb.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bersuker K, Olzmann JA. Establishing the lipid droplet proteome: Mechanisms of lipid droplet protein targeting and degradation. Biochim Biophys Acta. 2017;1862:1166–1177. doi: 10.1016/j.bbalip.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujimoto T, Parton RG. Not just fat: The structure and function of the lipid droplet. Cold Spring Harb Perspect Biol. 2011;3:a004838. doi: 10.1101/cshperspect.a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brasaemle DL, Wolins NE. Isolation of lipid droplets from cells by density gradient centrifugation. Curr Protoc Cell Biol. 2016;72:3.15.1–3.15.13. doi: 10.1002/cpcb.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zehmer JK, et al. Targeting sequences of UBXD8 and AAM-B reveal that the ER has a direct role in the emergence and regression of lipid droplets. J Cell Sci. 2009;122:3694–3702. doi: 10.1242/jcs.054700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olzmann JA, Richter CM, Kopito RR. Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proc Natl Acad Sci USA. 2013;110:1345–1350. doi: 10.1073/pnas.1213738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki M, et al. ELMOD2 is anchored to lipid droplets by palmitoylation and regulates adipocyte triglyceride lipase recruitment. Mol Biol Cell. 2015;26:2333–2342. doi: 10.1091/mbc.E14-11-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi K, et al. The lack of the C-terminal domain of adipose triglyceride lipase causes neutral lipid storage disease through impaired interactions with lipid droplets. J Clin Endocrinol Metab. 2008;93:2877–2884. doi: 10.1210/jc.2007-2247. [DOI] [PubMed] [Google Scholar]
- 14.Alexandru G, et al. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruber A, et al. The N-terminal region of comparative gene identification-58 (CGI-58) is important for lipid droplet binding and activation of adipose triglyceride lipase. J Biol Chem. 2010;285:12289–12298. doi: 10.1074/jbc.M109.064469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stevanovic A, Thiele C. Monotopic topology is required for lipid droplet targeting of ancient ubiquitous protein 1. J Lipid Res. 2013;54:503–513. doi: 10.1194/jlr.M033852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bussell R, Jr, Eliezer D. A structural and functional role for 11-mer repeats in alpha-synuclein and other exchangeable lipid binding proteins. J Mol Biol. 2003;329:763–778. doi: 10.1016/s0022-2836(03)00520-5. [DOI] [PubMed] [Google Scholar]
- 18.Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta. 2009;1791:419–440. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuberth C, Buchberger A. UBX domain proteins: Major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci. 2008;65:2360–2371. doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boeszoermenyi A, et al. Structure of a CGI-58 motif provides the molecular basis of lipid droplet anchoring. J Biol Chem. 2015;290:26361–26372. doi: 10.1074/jbc.M115.682203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson A, et al. Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904, and erratum (2006) 78:4235. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 22.Brown DA. Interactions between GPI-anchored proteins and membrane lipids. Trends Cell Biol. 1992;2:338–343. [PubMed] [Google Scholar]
- 23.Brunsveld L, et al. Lipidated ras and rab peptides and proteins—Synthesis, structure, and function. Angew Chem Int Ed Engl. 2006;45:6622–6646. doi: 10.1002/anie.200600855. [DOI] [PubMed] [Google Scholar]
- 24.Viklund H, Bernsel A, Skwark M, Elofsson A. SPOCTOPUS: A combined predictor of signal peptides and membrane protein topology. Bioinformatics. 2008;24:2928–2929. doi: 10.1093/bioinformatics/btn550. [DOI] [PubMed] [Google Scholar]
- 25.Schrul B, Kopito RR. Peroxin-dependent targeting of a lipid-droplet-destined membrane protein to ER subdomains. Nat Cell Biol. 2016;18:740–751. doi: 10.1038/ncb3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delaney E, Khanna P, Tu L, Robinson JM, Deutsch C. Determinants of pore folding in potassium channel biogenesis. Proc Natl Acad Sci USA. 2014;111:4620–4625. doi: 10.1073/pnas.1324274111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang J, et al. Characterization of erasin (UBXD2): A new ER protein that promotes ER-associated protein degradation. J Cell Sci. 2006;119:4011–4024. doi: 10.1242/jcs.03163. [DOI] [PubMed] [Google Scholar]
- 28.Schweiger M, Lass A, Zimmermann R, Eichmann TO, Zechner R. Neutral lipid storage disease: Genetic disorders caused by mutations in adipose triglyceride lipase/PNPLA2 or CGI-58/ABHD5. Am J Physiol Endocrinol Metab. 2009;297:E289–E296. doi: 10.1152/ajpendo.00099.2009. [DOI] [PubMed] [Google Scholar]
- 29.Poppelreuther M, et al. The N-terminal region of acyl-CoA synthetase 3 is essential for both the localization on lipid droplets and the function in fatty acid uptake. J Lipid Res. 2012;53:888–900. doi: 10.1194/jlr.M024562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horiguchi Y, Araki M, Motojima K. Identification and characterization of the ER/lipid droplet-targeting sequence in 17beta-hydroxysteroid dehydrogenase type 11. Arch Biochem Biophys. 2008;479:121–130. doi: 10.1016/j.abb.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 31.Kaczocha M, Glaser ST, Chae J, Brown DA, Deutsch DG. Lipid droplets are novel sites of N-acylethanolamine inactivation by fatty acid amide hydrolase-2. J Biol Chem. 2010;285:2796–2806. doi: 10.1074/jbc.M109.058461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang W, Napoli JL. The retinol dehydrogenase Rdh10 localizes to lipid droplets during acyl ester biosynthesis. J Biol Chem. 2013;288:589–597. doi: 10.1074/jbc.M112.402883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zehmer JK, Bartz R, Liu P, Anderson RG. Identification of a novel N-terminal hydrophobic sequence that targets proteins to lipid droplets. J Cell Sci. 2008;121:1852–1860. doi: 10.1242/jcs.012013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deisenroth C, Itahana Y, Tollini L, Jin A, Zhang Y. p53-Inducible DHRS3 is an endoplasmic reticulum protein associated with lipid droplet accumulation. J Biol Chem. 2011;286:28343–28356. doi: 10.1074/jbc.M111.254227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Howe V, Chua NK, Stevenson J, Brown AJ. The regulatory domain of squalene monooxygenase contains a re-entrant loop and senses cholesterol via a conformational change. J Biol Chem. 2015;290:27533–27544. doi: 10.1074/jbc.M115.675181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Granseth E, von Heijne G, Elofsson A. A study of the membrane-water interface region of membrane proteins. J Mol Biol. 2005;346:377–385. doi: 10.1016/j.jmb.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 38.Das R, et al. Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol Cell. 2009;34:674–685. doi: 10.1016/j.molcel.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bersuker K, et al. A proximity labeling strategy provides insights into the composition and dynamics of lipid droplet proteomes. Dev Cell. 2018;44:97–112.e7. doi: 10.1016/j.devcel.2017.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu F, Stanton JJ, Wu Z, Piwnica-Worms H. The human Myt1 kinase preferentially phosphorylates Cdc2 on threonine 14 and localizes to the endoplasmic reticulum and Golgi complex. Mol Cell Biol. 1997;17:571–583. doi: 10.1128/mcb.17.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Annis MG, et al. Bax forms multispanning monomers that oligomerize to permeabilize membranes during apoptosis. EMBO J. 2005;24:2096–2103. doi: 10.1038/sj.emboj.7600675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westphal D, et al. Apoptotic pore formation is associated with in-plane insertion of Bak or Bax central helices into the mitochondrial outer membrane. Proc Natl Acad Sci USA. 2014;111:E4076–E4085. doi: 10.1073/pnas.1415142111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pidoux G, et al. Optic atrophy 1 is an A-kinase anchoring protein on lipid droplets that mediates adrenergic control of lipolysis. EMBO J. 2011;30:4371–4386. doi: 10.1038/emboj.2011.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z, et al. Lipid droplets control the maternal histone supply of Drosophila embryos. Curr Biol. 2012;22:2104–2113. doi: 10.1016/j.cub.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krahmer N, et al. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 2011;14:504–515. doi: 10.1016/j.cmet.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barneda D, et al. The brown adipocyte protein CIDEA promotes lipid droplet fusion via a phosphatidic acid-binding amphipathic helix. eLife. 2015;4:e07485. doi: 10.7554/eLife.07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kory N, Thiam AR, Farese RV, Jr, Walther TC. Protein crowding is a determinant of lipid droplet protein composition. Dev Cell. 2015;34:351–363. doi: 10.1016/j.devcel.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eisenberg D, Weiss RM, Terwilliger TC. The helical hydrophobic moment: A measure of the amphiphilicity of a helix. Nature. 1982;299:371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- 49.Cornell RB. Membrane lipid compositional sensing by the inducible amphipathic helix of CCT. Biochim Biophys Acta. 2016;1861:847–861. doi: 10.1016/j.bbalip.2015.12.022. [DOI] [PubMed] [Google Scholar]
- 50.Gautier R, Douguet D, Antonny B, Drin G. HELIQUEST: A web server to screen sequences with specific alpha-helical properties. Bioinformatics. 2008;24:2101–2102. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- 51.Orgel JP. Surface-active helices in transmembrane proteins. Curr Protein Pept Sci. 2006;7:553–560. doi: 10.2174/138920306779025666. [DOI] [PubMed] [Google Scholar]
- 52.Wang CW, Lee SC. The ubiquitin-like (UBX)-domain-containing protein Ubx2/Ubxd8 regulates lipid droplet homeostasis. J Cell Sci. 2012;125:2930–2939. doi: 10.1242/jcs.100230. [DOI] [PubMed] [Google Scholar]
- 53.McAlister GC, et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem. 2014;86:7150–7158. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 55.Huttlin EL, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun LP, Li L, Goldstein JL, Brown MS. Insig required for sterol-mediated inhibition of Scap/SREBP binding to COPII proteins in vitro. J Biol Chem. 2005;280:26483–26490. doi: 10.1074/jbc.M504041200. [DOI] [PubMed] [Google Scholar]
- 57.Zhang L, Elias JE. Relative protein quantification using tandem mass tag mass spectrometry. Methods Mol Biol. 2017;1550:185–198. doi: 10.1007/978-1-4939-6747-6_14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.