Fig 1. Whi2 is required to suppress TORC1 activity in low amino acids.
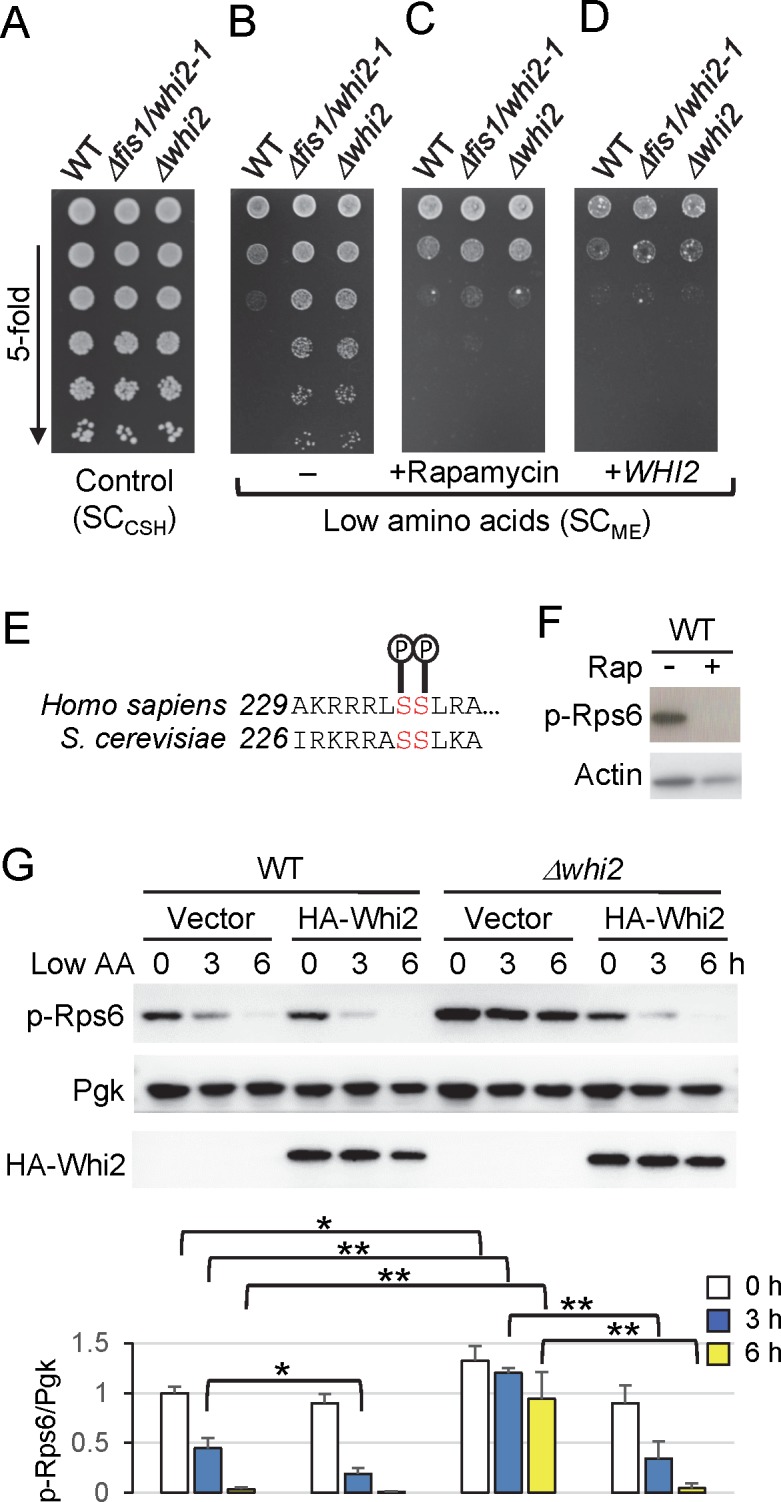
(A) Density-matched liquid yeast cultures grown in YPD were spotted on SCCSH (1,762 mg/L amino acids) agar plates in 5-fold serial dilutions (starting with undiluted samples) and incubated for 2 days. (B) The same cultures from panel A were spotted on SCME (1,200 mg/L amino acids) in the same manner and incubated for 3 days. (C) The same cultures from panel A were spotted on SCME containing 2.5 ng/mL rapamycin (Sigma). (D) Parallel cultures of yeast strains transformed with a WHI2 (CEN-URA) expression plasmid containing a genomic DNA fragment of the WHI2 ORF and its native regulatory sequences (Table 2) were analyzed as in panel B. Images in panels A-D were adjusted equally to reflect original results. Representative of many independent experiments are shown. (E) Amino acid sequence alignment of conserved phosphorylation sites near or at the C-terminus of human and yeast ribosomal protein S6, respectively. (F) Immunoblot of whole cell lysates prepared from wild type yeast treated with/without 200 nM rapamycin for 1 h in YPD and analyzed using anti-mammalian phospho-S6 (Ser235/S236) and actin antibodies. (G) TORC1 activity after switching from control/high (SCCSH) to low amino acid media (SCME) using anti-phospho-Rps6 immunoblots of lysates from strains transformed with empty vector or a constitutive PGK1 promoter-driven N-terminal HA-tagged Whi2 expression vector. Equal loading of samples was achieved primarily by using density-matched cultures and monitored with anti-Pgk. Corresponding bars below show quantification of TORC1 activity (the ratio of phospho-Rps6/Pgk). Values were normalized to the average value of WT transformed with empty vector at time zero, and presented in the bar graph as means ± SD (n = 4 independent experiments). **P < 0.01; *P < 0.05; using a two-tailed T test.
