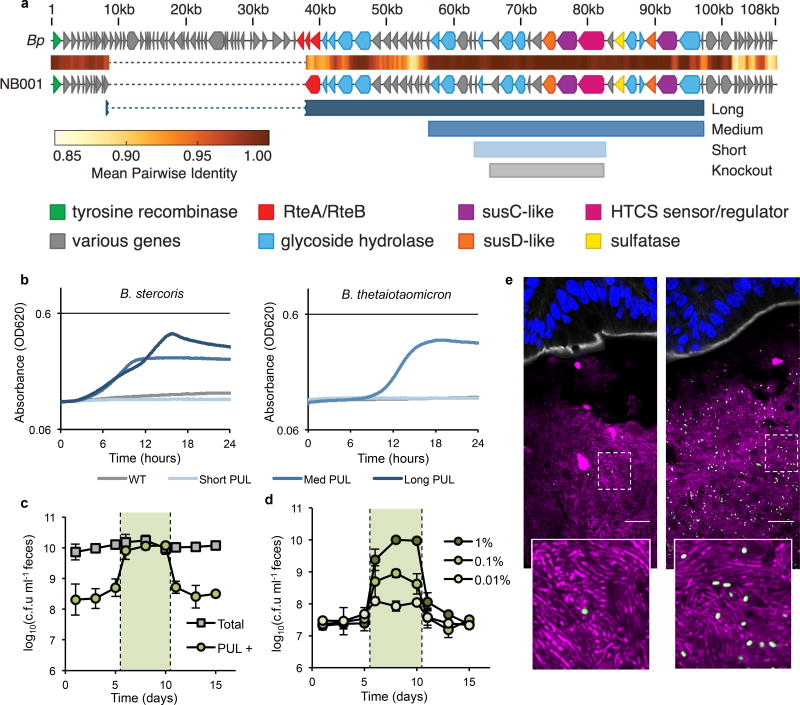
Figure 3. Control over population size can be engineered, and is highly tunable.
a, Schematic of porphyran PUL from NB001 aligned to the previously published B. plebeius PUL. Shown below are the different minimal PULs (Long = 34 genes, Medium = 21 genes, and Short = 10 genes) designed and tested for ability to confer growth on porphyran. The eight gene region deleted from NB001 PUL− is shown in gray (Knockout). b, Growth curves for natural and engineered strains on porphyran as sole carbon source. c, B. thetaiotaomicron harboring the medium length PUL colonized in conventional mice consuming a MAC-rich diet demonstrates toggling upon administration of 1% w/v porphryan in the drinking water (shaded green, n=4). Error bars indicate standard deviation. d, NB001 PUL+ colonized in conventional mice consuming a MAC-rich diet demonstrates tunable (finely controlled) response to porphyran in the drinking water (shaded green, n=4 per group). Error bars indicate standard deviation. e, NB001 expressing GFP colonized in conventional mice given 0.01% porphryan (left) or 1% porphyran (right). Image is of proximal colon with host epithelium visualized by DAPI (nuclei, blue), epithelial border visualized by phalloidin (F-actin, white), background microbiota by DAPI segmented from host epithelium (bacteria, magenta), NB001 by endogenous GFP (bacteria, green). Scale bar represents 16 µm.