Abstract
Base editors enable targeted single-nucleotide conversion in genomic DNA. Here we show that expression levels are a bottleneck in base editing efficiency. We optimize cytidine (BE4) and adenine (ABE7.10) base editors by modification of nuclear localization signals and codon usage, and ancestral reconstruction of the deaminase component. The resulting BE4max, AncBE4max, and ABEmax editors correct pathogenic SNPs with substantially increased efficiency in a variety of mammalian cell types.
Point mutations represent the majority of known pathogenic human genetic variants1. Base editors enable the direct installation and correction of targeted point mutations in genomic DNA. These fusion proteins include a catalytically impaired Cas9, natural or laboratory-evolved nucleobase deaminases, and, in some cases, proteins that help preserve the resulting single-nucleotide change2,3. Cytidine base editors (e.g., BE44), convert target C:G base pairs to T:A and adenine base editors (e.g., ABE7.103) convert A:T to G:C. Collectively, these editors enable the targeted installation of all four transition mutations (C-to-T, G-to-A, A-to-G, and T-to-C), which account for 61% of human pathogenic SNPs in the ClinVar database (Supplementary Fig. 1a, 1b). Base editors have been successfully used in diverse systems including prokaryotes, plants, fish, amphibians, mammals, and human embryos4–8. However, diminished efficiency of base editors at certain target sites or in particular cell types limits their utility.
To test if base editing in cells is limited by transfection efficiency of base editor plasmid or by base editor expression, we transfected HEK293T cells with three-plasmid mixtures in which one plasmid expresses mCherry (transfection marker), another expresses a targeting sgRNA, and a third expresses either (1) BE4 alone, (2) BE4 and GFP on separate promoters to follow transfection of this plasmid, or (3) a BE4–P2A–GFP fusion to directly follow BE4 expression (Fig. 1a). P2A is a self-cleaving peptide9 that couples GFP production with full-length BE4 production.
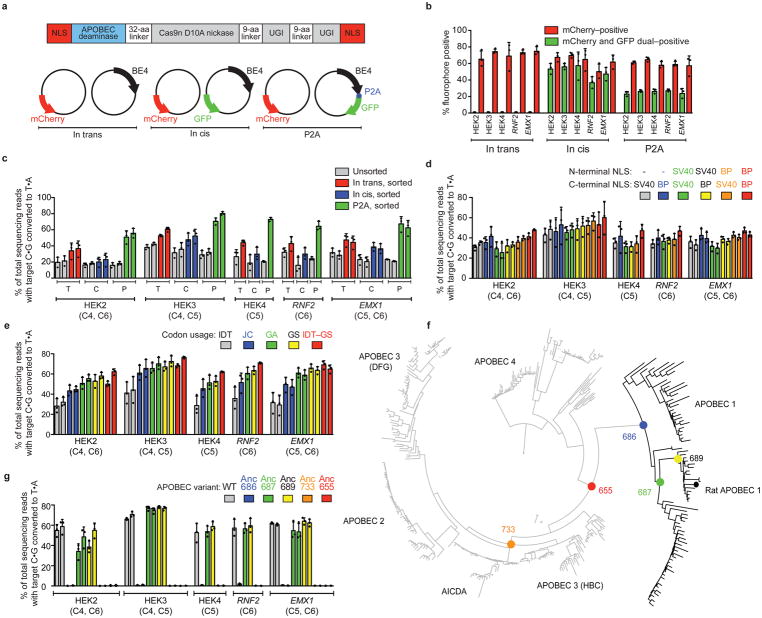
Figure 1. Identifying and addressing factors that limit base editing efficiency in mammalian cells.
(a) Plasmids used to elucidate the relationship between base editor expression and editing efficiency in mammalian cells: mCherry (transfection control), and either BE4 (“in trans”), BE4 and GFP on separate promoters (“in cis”), or BE4–P2A–GFP (“P2A”). (b) Percent mCherry-positive or GFP-positive HEK293T cells 3 days after transfection of the constructs in (a). (c) Target C:G-to-T:A editing in unsorted and sorted HEK293T cells. Sorted in trans cells were mCherry-positive, while sorted in cis and P2A cells were mCherry and GFP double-positive. (d) Effects of six NLS configurations on BE4 editing efficiency at five genomic loci in HEK293T cells. (e) Effects of five codon usages on editing efficiency of bis-bpNLS-BE4 in HEK293T cells. IDT=Integrated DNA Technologies; JC=Jeff Coller; GA=GeneArt; GS=GenScript; IDT-GS=IDT APOBEC+GenScript Cas9 nickase. (f) Phylogenetic tree for ancestral APOBEC reconstruction. (g) Base editing of bis-bpNLS-BE4 variants with GenScript codons using the ancestral APOBEC domains in (f) in HEK293T cells. Values and error bars represent the mean and standard deviation of n=3 biologically independent experiments (dots) 3 days after transfection.
Transfection with (1) and collecting mCherry-positive cells resulted in 45±7.1% average C:G-to-T:A conversion within the base editing activity window (positions 4–8, counting the PAM as positions 21–23) at five genomic loci (Fig. 1b, c). The average editing efficiency among mCherry and GFP double-positive cells did not improve with (2) (Fig. 1c), suggesting that transfection efficiency was not limiting editing efficiency. In contrast, double-positive cells following transfection with (3) exhibited 65±6.4% editing, 1.9-fold higher than in sorted cells following (2) (Fig. 1c), indicating that cells expressing base editors and/or the amount of functional editor protein produced by each cell are major bottlenecks of editing efficiency.
To optimize nuclear localization, we tested all six combinations of BE4 N- and C-terminal fusions to the SV40 NLS used in BE4, or to a bipartite NLS (bpNLS) (Fig. 1d, Supplementary Fig. 2)10. A bpNLS at both the N- and C-termini (bis-bpNLS) performed best, resulting in a 1.3-fold average improvement in BE4-mediated C:G-to-T:A editing efficiency at five genomic loci (Fig. 1d, Supplementary Fig. 2a).
Next we generated bis-bpNLS BE4 variants using eight codon usages: from IDT (used in BE44), GeneArt, Coller and co-workers11, and GenScript. Every codon optimization method improved editing efficiency over IDT codon usage in HEK293T cells (Fig. 1e, Supplementary Fig. 2b). We also tested four chimeric codon-optimized BE4 variants that mixed different deaminase and Cas9 nickase codon usages (Supplementary Fig. 3a, 3b), but none outperformed the GenScript-only variant (BE4max), which induced 1.8-fold higher editing over bis-bpNLS BE4 with IDT codons (Fig. 1e).
Chimeric editor experiments implicated expression of the APOBEC1 cytidine deaminase and Cas9 nickase as determinants of base editing efficiency (Fig. 1e and Supplementary Fig. 3a). To further enhance APOBEC1 expression, we performed ancestral sequence reconstruction (ASR) using 468 APOBEC homologs (Supplementary File 1, Supplementary Sequences 1). ASR uses an alignment of protein sequences, an evolutionary model, and a resulting phylogenetic tree to infer ancestral sequences12, and can improve protein expression while preserving activity13,14. We created a maximum-likelihood APOBEC phylogeny and inferred the most likely sequences at ancestral nodes (Fig. 1f), then constructed five GenScript-coded bis-bpNLS-BE4 variants from five ancestral cytidine deaminases (Supplementary Fig. 4). Two ancestors, Anc689 and Anc687, containing 36 and 45 amino acid substitutions relative to rAPOBEC1, respectively, resulted in bis-bpNLS-BE4 variants that efficiently edited five test loci in HEK293T cells (Fig. 1g).
To characterize the base editing activities of these optimized variants under sub-optimal conditions, we compared eight different plasmid doses of BE4, BE4max, and AncBE4max (bis-bpNLS BE4 with the Anc689 APOBEC and GenScript codons) at three genomic loci in HEK293T cells (Fig. 2a). AncBE4max showed the highest activity across all tested sites over a range of plasmid doses, with BE4max performing slightly below or similar to, those of AncBE4max (Fig. 2a, Supplementary Fig. 5). AncBE4max and BE4max improved over BE4 at rates ranging from 1.7-fold at higher plasmid doses to >9-fold at lower plasmid doses (Fig. 2a). Product purities of BE4max and AncBE4max—ratios of desired point mutations to indels and undesired mutations at the target nucleotide—were better than or comparable to those of BE4 (Supplementary Fig. 6, 7a). The shape of the base editing activity window for BE4max and AncBE4max was unchanged compared to that of BE4 (Supplementary Fig. 8a).
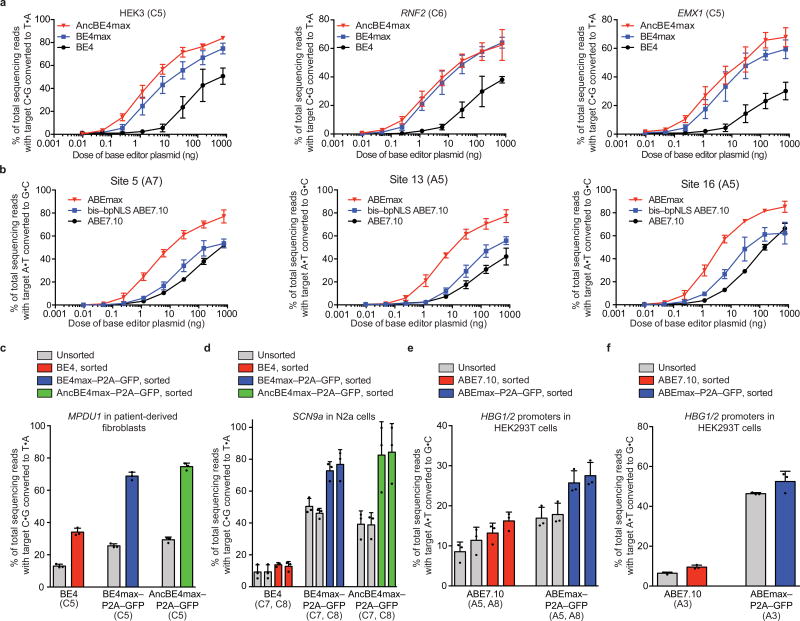
Figure 2. Properties of optimized AncBE4max, BE4max, and ABEmax compared to BE4 and ABE7.10.
(a) Comparison at three genomic loci in HEK293T cells across eight plasmid doses. (b) Comparison of ABE7.10, bis-bpNLS-ABE with IDT codons, and ABEmax at three genomic loci in HEK293T cells across eight plasmid doses. (c) C:G-to-T:A editing for the correction of MPDU1 Leu119Pro by BE4, BE4max, or AncBE4max in unsorted or sorted fibroblasts derived from congenital disorder of glycosylation (CDG) type 1f patients. BE4 samples were sorted for mCherry and BE4max–P2A–GFP and AncBE4max–P2A–GFP samples for GFP-positive cells. (d) C:G-to-T:A editing of the 3′ splice acceptor in intron 6 of SCN9a in mouse N2a neuroblastoma cells, unsorted or sorted as described in (c). (e) A:T-to-G:C editing for the installation of -116 A>G and -113 A>G in the HBG fetal hemoglobin promoters by ABE7.10 or ABEmax in HEK293T cells. ABE samples were sorted for mCherry-positive cells, and ABEmax–P2A–GFP samples were sorted for mCherry and GFP double-positive cells. (f) A:T-to-G:C editing to install HBG promoter mutation -175 T>C by ABE7.10 or ABEmax in HEK293T cells, unsorted or sorted as described in (e). Values and error bars represent the mean and standard deviation of n=3 biologically independent experiments (dots) 3 days after transfection.
BE4max and AncBE4 max showed >3-fold and >5-fold higher mRNA (Supplementary Fig. 9a) and protein (Supplementary Fig. 9b) expression in HEK293T cells relative to BE4, improvements that correlated with editing efficiency. Among transfectable HEK293T cells expressing BE4max–P2A–GFP and AncBE4max–P2A–GFP (mCherry and GFP double-positive), base editing at three genomic loci averaged 89±0.9% and 90±1.5%, respectively, while double-positive cells expressing BE4–P2A–GFP averaged 48±8.0% (Figure 1e, Supplementary Fig. 9c, d). Thus isolating cells expressing BE4max and AncBE4max results in much higher editing frequencies, which could facilitate creation of cell lines, agricultural strains, or animal models.
Adenine base editors (ABEs) use a laboratory-evolved deoxyadenosine deaminase and a Cas9 nickase to mediate the conversion of target A:T to G:C base pairs3, reversing the most common class of point mutations in living systems (Supplementary Fig. 1b)15. We applied the above improvements to ABE7.103. Replacing the SV40 NLS in ABE7.10 with the bis-bpNLS increased editing efficiencies ~1.5- to 2-fold at sub-optimal ABE doses in HEK293T cells (Fig. 2b). GenScript codon optimization of bis-bpNLS ABE7.10 (ABEmax) resulted in 1.3- to 7.9-fold higher editing levels than IDT codon usage (in ABE7.10) at high and low plasmid doses, respectively (Fig. 2b, Supplementary Fig. 10, 11). Although indels from ABEmax remained rare (≤1.6%), they were elevated from the virtually undetectable indel levels of ABE7.103 (Supplementary Fig. 12). ABEmax exhibited increases in mRNA and protein levels in HEK293T cells compared to ABE (Supplementary Fig. 13), and product purity and the editing window remained unchanged (Supplementary Fig. 7b, Supplementary Fig. 8b). These findings establish that improvements in nuclear localization and codon usage that benefit BE4 also enhance ABE efficiency.
We evaluated the ability of BE4max, AncBE4max, and ABEmax (Supplementary Sequences 2, 3) to edit disease-relevant loci in diverse cell types. Patient-derived fibroblasts harboring the MPDU1 Leu119Pro T>C mutation that drives congenital disorder of glycosylation type 1f16 were nucleofected with plasmids expressing BE4, BE4max–P2A–GFP, or AncBE4max–P2A–GFP. The pathogenic SNP was corrected 2.0- and 2.2-fold more efficiently by BE4max (26±1.3% unsorted, 69±2.5% sorted) and AncBE4max (29±1.7% unsorted, 75±2.2% sorted) than by BE4 (13±1.2% unsorted, 34±2.4% sorted) (Fig. 2c, Supplementary Fig. 14a). Second, we used BE4max and AncBE4max to mutate the splice acceptor of SCN9a intron 6a in mouse N2a neuroblastoma cells in the chronic pain-associated voltage-gated sodium channel NaV1.7 (SCN9a gene)17. BE4, BE4max and AncBE4max respectively resulted in 9.3±4.4%, 50±5.0% and 39±7.7% (unsorted) or 14±1.3%, 77±9.3% and 84±18% (sorted) editing (Fig. 2d, Supplementary Fig. 14a), 4.2- to 6.0-fold improvements favoring BE4max and AncBE4max. In one sorted sample, 99.8% of cells expressing AncBE4max contained mutations at both targeted SCN9a nucleotides (Supplementary Fig. 15). Third, we used ABEmax to install activating mutations in the promoters of HBG1 or HBG2 (γ-globin) that can rescue β-globin disorders with two sgRNAs: (1) -116 A>G and -113 A>G; and (2) -175 T>C18,19. For the first sgRNA, ABEmax resulted in approximately double the -116 A>G and -113 A>G conversion than ABE7.10 in both unsorted and sorted HEK293T cells (Fig. 2e). For the second sgRNA, ABE7.10 and ABEmax respectively induced 6.5±0.57% and 46±0.55% (unsorted) and 10±1.0% and 52±5.2% (sorted) editing in HEK293T cells (Fig. 2f, Supplementary Fig. 14c), representing 5.2- and 7.1-fold improvements favoring ABEmax.
BE4max, AncBE4max, and ABEmax thus offer increased editing in a variety of settings, especially under sub-optimal conditions or at sites previously edited with modest efficiency. These improvements in expression and nuclear localization should also benefit other base editor delivery methodologies, including viral, mRNA, and RNP delivery.
Methods
General Methods
PCR was performed using either Phusion U Green Multiplex PCR Master Mix (ThermoFisher Scientific) or Q5 Host Start High-Fidelity 2x Master Mix (New England Biolabs) unless otherwise noted. All plasmids were assembled by either the USER cloning method as previously described20 or by Gibson assembly21. Plasmids for mammalian cell transfections were prepared using an endotoxin removal plasmid purification system, ZymoPURE Plasmid Midiprep (Zymo Research Corporation).
Cell culture conditions
HEK293T cells (ATCC CRL-3216) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Corning) supplemented with 10% fetal bovine serum (FBS), penicillin, and streptomycin (ThermoFisher Scientific). Fibroblast cell lines were maintained in DMEM supplemented with 15% FBS. N2a cells were maintained in DMEM supplemented with 10% FBS.
HEK293T transfection and genomic DNA preparation
HEK293T cells were seeded into 48-well Poly-D-Lysine coated plates (Corning) in the absence of antibiotic. 12–15 hours after plating, cells were transfected with 1 μL of Lipofectamine 2000 (ThermoFisher Scientific) using 750 ng of base editor plasmid, 250 ng of guide RNA plasmid, and 20 ng of fluorescent protein expression plasmid as a transfection control. Unless otherwise stated, cells were cultured for 3 days before they were washed with PBS (ThermoFisher Scientific). Genomic DNA was extracted by addition of 150 μL of freshly prepared lysis buffer (10 mM Tris-HCl, pH 7.5, 0.05% SDS, 25 μg/mL proteinase K (ThermoFisher Scientific)) directly into each transfected well. The resulting mixture was incubated for 1 hour at 37 °C before a 30-min enzyme inactivation step at 80 °C. Guide RNA sequences for HEK2, HEK3, HEK4, RNF2, EMX1, Site 2, Site 5, Site 13, Site 16 were previously reported.2–4 SCN, MPDU1, HBG Site 1, and HBG Site 2 were cloned as described in Supplementary Sequences 4.
HEK293T base editing dose titrations
HEK293T cells were seeded as described above and transfected with a mixture of base editor plasmid, guide RNA plasmid, pUC, and GFP. 250 ng of guide RNA plasmid and 20 ng of GFP transfection control plasmid were used for all samples. Base editor and pUC plasmids were combined in different amounts to maintain a constant amount of total DNA per transfection.
Fluorescence-activated cell sorting
Flow cytometry analysis was carried out using a FACSAria II (BD Biosciences). HEK293T cells were transfected with guide RNA expression plasmids, fluorophore expression plasmids, and editor expression plasmids. In trans samples were sorted for mCherry-positive cells. Both the in cis and P2A samples were sorted for both GFP and mCherry double-positive cells. A stringent mCherry-positive gate was used to avoid mCherry false positives. N2a cells and fibroblasts were sorted for mCherry-positive and GFP-positive cells. Genomic DNA for sorted and unsorted FACS samples was isolated using the Agencourt DNAdvance Genomic DNA Isolation Kit (Beckman Coulter) according to the manufacturer’s instructions. Gating for all cell types can be found in Supplementary File 2.
Nucleofection of fibroblasts and genomic DNA extraction
Cells were nucleofected using the Primary P2 Cell Line 4D-Nucleofector X Kit (Lonza) according to manufacturer’s protocol. 1.25×105 cells were nucleofected in 20 μL of P2 buffer supplemented with 750 ng of editor, 250 ng of guide RNA plasmid, and 20 ng of mCherry nucleofection marker. Cells were nucleofected in a 16-well nucleocuvette strip using the DT-130 program. Following a 3-day incubation, cells were flow sorted and genomic DNA was extracted as described for HEK293T cells above.
High-throughput DNA sequencing (HTS) of genomic DNA
HTS of genomic DNA from HEK293T cells was performed as described previously2–4. For fibroblasts, 34 cycles of amplification were used for PCR1. Primers for PCR 1 of HEK2, HEK3, HEK4, RNF2, EMX1, ABE Site 2, ABE Site 5, ABE Site 13, ABE Site 16, and HBG loci were used as previously described3,4,22. PCR 1 primers for type 1F congenital glycosylation disorder, SCN9a, and all previously used loci are listed in Supplementary Sequences 5.
General HTS analysis
Sequencing reads were demultiplexed using the MiSeq Reporter (Illumina) and Fastq files were analyzed using using open source analysis tools. FASTQ files were aligned to the reference genome using the burrows-wheeler aligner (bwa-mem)23. Statistics for each base were calculated using the pysamstats utility available at https://github.com/alimanfoo/pysamstats. All reads for a given base were aligned to the reference sequence. Total reads were the sum of all base calls, insertions, and deletions at any given nucleotide position. Percent representation of each base was calculated as reads of a given base divided by total reads. Indel frequencies were quantified with a custom Matlab script as previously described3,24.
Quantitative RT-PCR and quantitative PCR
HEK293T cells were transfected with base editor–P2A–GFP plasmids and incubated 3 days before harvesting DNA and RNA from each sample. DNA samples were harvested using the genomic DNA preparation protocol described above. RNA was isolated and amplified using the Cells-to-Ct (ThermoFisher) kit according to the manufacturer’s protocol except the DNase treatment step used 2X DNase for twice as long to ensure complete degradation of plasmid DNA. Levels of mRNA were calculated by normalizing base editor mRNA levels to β-actin levels by ΔΔCt. Plasmid DNA levels, as determined by qPCR of the BGH poly-adenylation sequence present on the base editor plasmid, were normalized to β-actin levels to ensure that mRNA abundance was not skewed by transfection efficiency.
Western blotting
HEK293T cells were transfected with 750 ng of base editor–3X HA tag plasmid and 250 ng of guide RNA plasmid. After 3 days, cells were lysed using RIPA buffer with PMSF and cOmplete Protease Inhibitor Cocktail (Roche). Samples were boiled and quantified using a BCA assay. 10 μg of protein was loaded per well into a 12-well 4–12% Tris gel (Novex), dry-transferred to nitrocellulose paper for 7 min at 20 V before blocking and incubation with anti-HA (Cell Signaling Technology) and anti-Actin antibodies (Cell Signaling Technology) and visualized using an Odyssey imager. Uncropped blots are shown in Supplementary Fig. 16.
APOBEC sequence collection
APOBEC protein sequences used in phylogenetic analyses were identified through searches of the UniProt database25 with the BLASTP algorithm26 using selected query sequences. All sequences from these searches that returned BLASTP E-values < 10−7 were downloaded from UniProt. To reduce phylogenetic complexity, sequences were curated based on character length and pairwise sequence identity within each dataset. The dataset used for the construction of the non-redundant phylogeny was generated using four query sequences: UniProt IDs P41238, H2P4E7, E1BTD6, and H2P4E9. Multiple sequences were necessary to generate full coverage due to the low sequence identity across the family, which is <25% between some members. Limits were chosen to remove truncated and partial sequences and those featuring large insertions or terminal extensions. Sequences greater than 97% identical, determined by pairwise alignment within the dataset, were also removed. This level of identity provides a high level of detail within the tree while accelerating computational time by removing redundant taxa. The final dataset contains 468 taxa (Supplementary Sequences 1).
Phylogeny construction
A multiple sequence alignment of the dataset was generated with the program MAFFT using the FFT-NS-I x1000 algorithm27 (Supplementary File 1). Model selection used the Bayesian information criteria (BIC) to determine the evolutionary model that best fit the input alignment28. 228 models where tested. The Jones Taylor Thornton (JTT) substitution matrix with empirical frequencies (F) and free rates with five categories (R5) was the model that best fit the data. A maximum likelihood (ML) phylogenetic tree was inferred with IQ-TREE29 using the best fit model (JTT+F+R5). The starting trees were generated by randomized maximum parsimony and searched by fast hill-climbing Nearest Neighbor Interchange (NNI). Tree topology, branch lengths, and rate parameters were optimized. Branch supports were estimated with Ultrafast boot strapping, implemented in IQ-TREE30 (Supplementary Fig. 17, Supplementary File 3).
Ancestral sequence reconstruction
Sequences at internal nodes in the phylogeny were inferred using the codeml program from the PAML software package31. Posterior amino acid probabilities at each site were calculated using the JTT substitution matrix, given the ML tree and estimated background frequencies generated by IQ-TREE. N- and C-termini of ancestral sequences were modified manually to match those of rat APOBEC1.
Statistics and reproducibility
All statistical analyses were performed on n=3 biologicaly independent experiments using unpaired Student’s t-test. Biologically independent experiments reported here are from independent splits of each cell type used. Degrees of freedom = 4.
Clinvar analysis
Custom code provided in Supplementary Note 1 was used to determine base pair changes required to correct pathogenic SNPs in the ClinVar database 1.
Life sciences reporting summary
Further information on experimental design and reagents is available in the Life Sciences Reporting Summary.
Supplementary Material
Acknowledgments
This work was supported by the Ono Pharma Foundation, DARPA HR0011-17-2-0049, U.S. NIH RM1 HG009490, R01 EB022376, and R35 GM118062, and HHMI. Flow cytometry was supported by NCI P30CCA14051. L.W.K. is an NSF Graduate Research Fellow and was supported NIH Training Grant T32 GM095450. J.L.D. gratefully acknowledges graduate fellowship support from the NSF and Hertz Foundation. We thank Jeff Coller, Gavin Hansen, Mitch Weiss, and Akshay Sharma for helpful discussions.
Footnotes
Author Contributions
L.W.K., J.L.D., C.W., J.M.L., T.T., G.A.N., and J.P.M. generated reagents and conducted experiments. C.W. and A.R. performed computational analyses. D.R.L supervised the research. All authors contributed to writing the manuscript.
Data Availability
Plasmids encoding BE4max, AncBE4max, and ABEmax have been deposited to Addgene. High-throughput sequencing data are deposited in the NCBI Sequence Read Archive (SRP145378).
Competing Financial Interests
The authors declare competing financial interests: D.R.L. is a consultant and co-founder of Editas Medicine, Pairwise Plants, and Beam Therapeutics, companies that use genome editing. L.W.K., J.L.D., C.W., and D.R.L. have filed patent applications on aspects on this work. The authors declare no competing non-financial interests.
References
- 1.Landrum MJ, et al. Nucleic acids research. 2016;44:D862–868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komor AC, et al. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaudelli NM, et al. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komor AC, et al. Sci Adv. 2017;3:eaao4774. doi: 10.1126/sciadv.aao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li G, et al. Protein Cell. 2017;8:776–779. doi: 10.1007/s13238-017-0458-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang P, et al. Protein Cell. 2017;8:811–822. doi: 10.1007/s13238-017-0475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryu SM, et al. Nature biotechnology. 2017 [Google Scholar]
- 8.Hess GT, Tycko J, Yao D, Bassik MC. Mol Cell. 2017;68:26–43. doi: 10.1016/j.molcel.2017.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JH, et al. PloS one. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki K, et al. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanson G, Coller J. Nature reviews Molecular cell biology. 2018;19:20–30. doi: 10.1038/nrm.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harms MJ, Thornton JW. Nature reviews Genetics. 2013;14:559–571. doi: 10.1038/nrg3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wheeler LC, Lim SA, Marqusee S, Harms MJ. Curr Opin Struct Biol. 2016;38:37–43. doi: 10.1016/j.sbi.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Risso VA, et al. J Am Chem Soc. 2013;135:2899–2902. doi: 10.1021/ja311630a. [DOI] [PubMed] [Google Scholar]
- 15.Krokan HE, Drablos F, Slupphaug G. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 16.Schenk B, et al. The Journal of clinical investigation. 2001;108:1687–1695. doi: 10.1172/JCI13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett DL, Woods CG. The Lancet Neurology. 2014;13:587–599. doi: 10.1016/S1474-4422(14)70024-9. [DOI] [PubMed] [Google Scholar]
- 18.Liu N, et al. Cell. 2018;173:430–442.e417. [Google Scholar]
- 19.Amato A, et al. International journal of laboratory hematology. 2014;36:13–19. doi: 10.1111/ijlh.12094. [DOI] [PubMed] [Google Scholar]
- 20.Badran AH, et al. Nature. 2016;533:58–63. doi: 10.1038/nature17938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibson DG, et al. Nature methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 22.Kim YB, et al. Nature biotechnology. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Durbin R. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu JH, et al. Nature. 2018;556:57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.UniProt Consortium, T. Nucleic acids research. 2018;46:2699. doi: 10.1093/nar/gky092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul SF, et al. Journal of molecular biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Katoh K, Standley DM. Molecular biology and evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalyaanamoorthy S, et al. Nature methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoang DT, et al. Mol Biol Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Z. Molecular biology and evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.