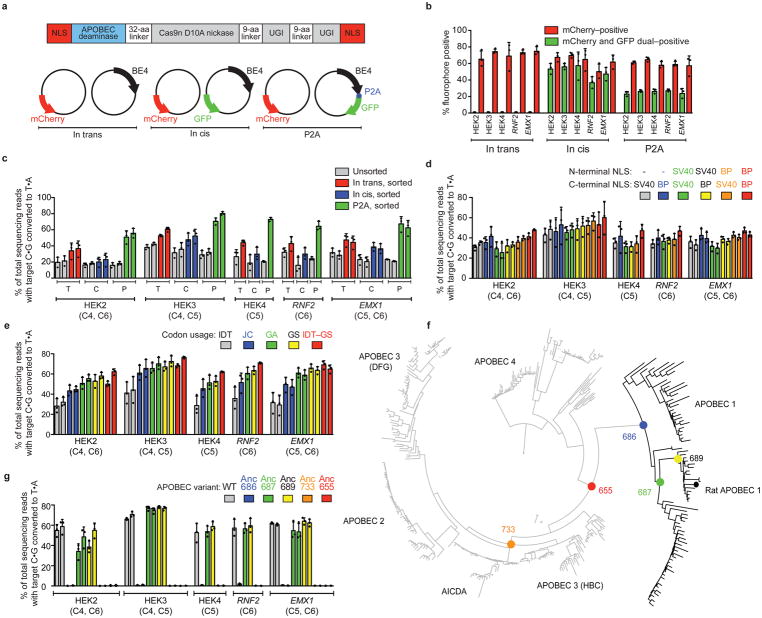
Figure 1. Identifying and addressing factors that limit base editing efficiency in mammalian cells.
(a) Plasmids used to elucidate the relationship between base editor expression and editing efficiency in mammalian cells: mCherry (transfection control), and either BE4 (“in trans”), BE4 and GFP on separate promoters (“in cis”), or BE4–P2A–GFP (“P2A”). (b) Percent mCherry-positive or GFP-positive HEK293T cells 3 days after transfection of the constructs in (a). (c) Target C:G-to-T:A editing in unsorted and sorted HEK293T cells. Sorted in trans cells were mCherry-positive, while sorted in cis and P2A cells were mCherry and GFP double-positive. (d) Effects of six NLS configurations on BE4 editing efficiency at five genomic loci in HEK293T cells. (e) Effects of five codon usages on editing efficiency of bis-bpNLS-BE4 in HEK293T cells. IDT=Integrated DNA Technologies; JC=Jeff Coller; GA=GeneArt; GS=GenScript; IDT-GS=IDT APOBEC+GenScript Cas9 nickase. (f) Phylogenetic tree for ancestral APOBEC reconstruction. (g) Base editing of bis-bpNLS-BE4 variants with GenScript codons using the ancestral APOBEC domains in (f) in HEK293T cells. Values and error bars represent the mean and standard deviation of n=3 biologically independent experiments (dots) 3 days after transfection.