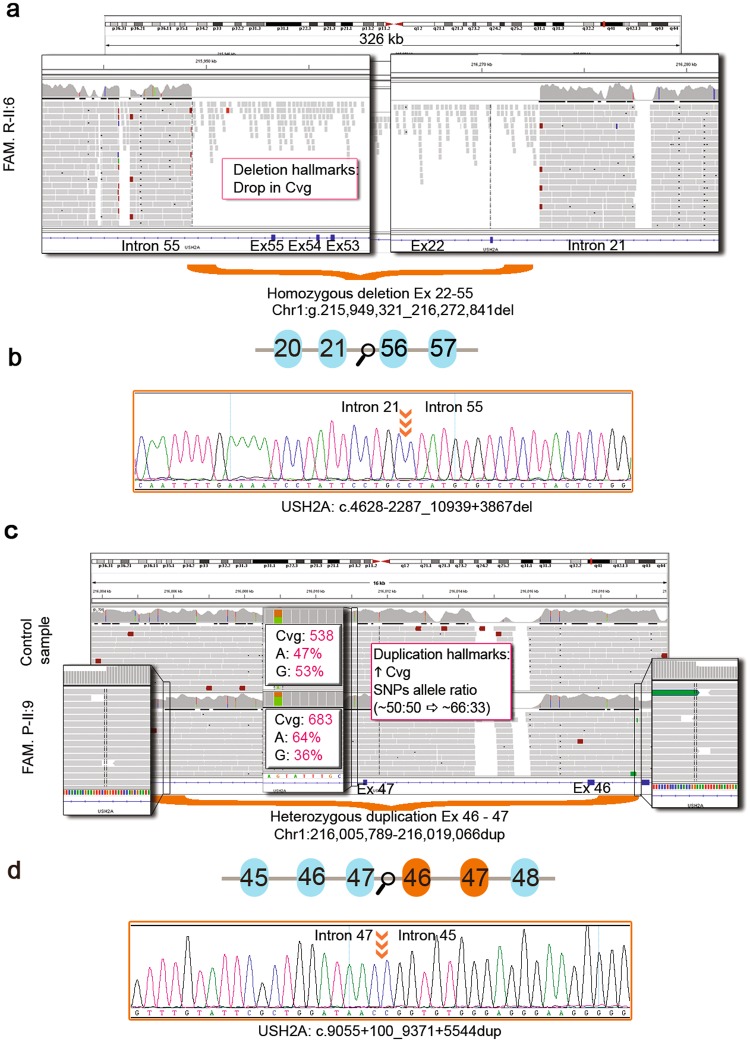
Figure 1.
Detection of CNVs in USH2A using our NGS approach. (a) IGV snapshot showing the homozygous deletion of exons 22–55 detected in family R-II:6 (Chr1:g.215,949,321_216,272,841del, hg19). The capture of whole genomic sequence of USH2A allowed us to determinate the CNVs breakpoints. (b) Schematic representation of the mutated gDNA sequence and Sanger sequencing of the breakpoint area (orange arrows) confirming the USH2A deletion (c.4628-2287_10939 + 3867del; NM_206933). (c) IGV snapshot showing the heterozygous duplication of exons 46–47 (Chr1:g.216,005,789_216,019,066dup, hg19) detected in family P-II:9 versus a control sample. The heterozygous duplication can be inspected visually using IGV paying special attention to (i) a sharp increase in the coverage and (ii) changes in the allele ratios of all the SNPs within the duplicated interval from ~50:50 to ~67:33 unmasking the presence of a total of three copies. (d) Schematic representation of the mutated gDNA sequence and Sanger sequencing of the breakpoint area (orange arrows) confirming the USH2A tandem duplication of exons 46 and 47 (c.9055 + 100_9371 + 5544dup; NM_206933).