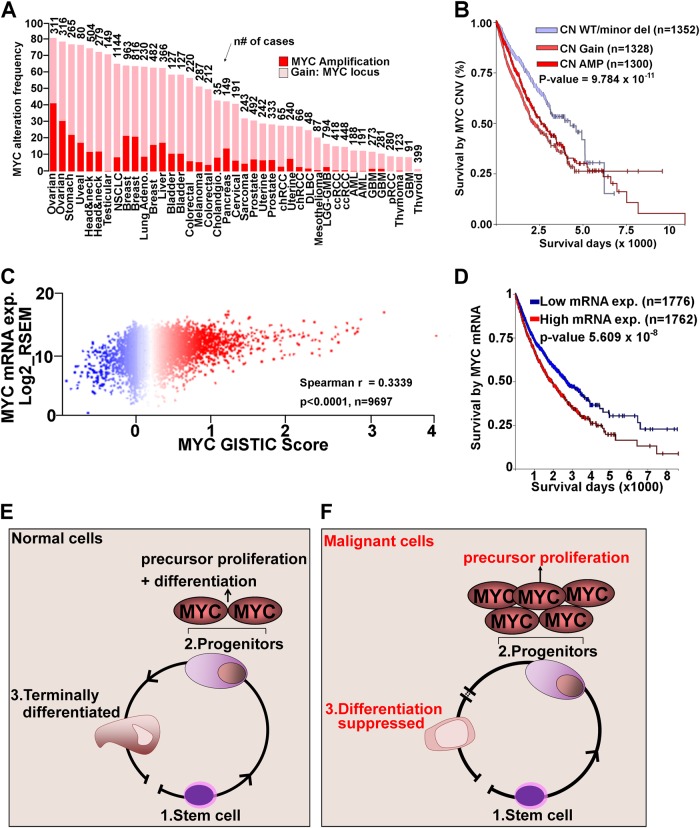
Fig. 1. MYC alterations across multiple human malignancies.
a TCGA and IGCG data were analyzed through cBioPortal to determine aberrations at the MYC locus using pre-assigned GISTIC scores in multiple cancers from different tissue types. b We analyzed TCGA PANCAN data sets available through TCGA hub in Xena Browser. Survival analysis of cases with copy number (CN) gains and amplification at the MYC loci vs. those with CN WT/minor deletion of MYC demonstrated a significant overall survival (p-value < 9.784E−11, LogRank test, n = 1352 WT/minor del, 2628 CN gain and amplification). Survival data analyzed in Xena Browser (https://xenabrowser.net/) c Anlysis of MYC GISTIC Score vs. MYC mRNA expression using PANCAN RNA-seq data available in TCGA hub in Xena Browser. There was a strong correlation with spearman r = 0.3339, p < 0.0001, n = 9697. d Survival analysis of patients with increased MYC mRNA compared to those with decreased MYC mRNA expression. Expression levels are normalized relative to expression levels in normal tissues. Increased MYC mRNA was associated with poor survival (n = 1762) compared to decreased MYC mRNA (n = 1776, p = 5.06 × 10−18 e Schematic representation of metazoan differentiation and how differentiation is stalled in malignant cells. Differentiation continuum is initiated through stem cells lineage commitment, followed by exponential proliferation of tissue precursors/progenitors mediated by two copies of the MYC gene. To maintain homeostasis, MYC-mediated proliferation is dominantly antagonized by terminal differentiation pathways. f Human malignancies have impaired differentiation that fails to antagonize the MYC gene allowing for exponential proliferation of tissue precursors