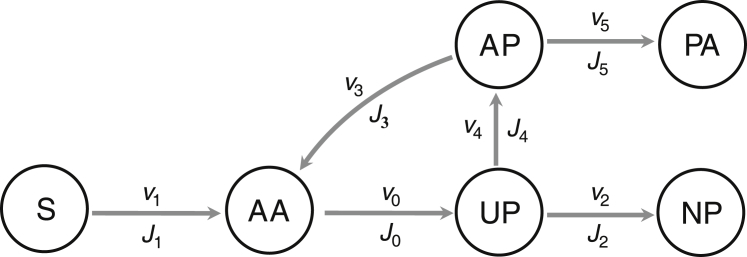
Figure 2.
Schematic illustration of protein resource fluxes in bacteria. AAs inside the cell come from the uptake and metabolism of the nutritional substrate (S) via P1 (flagella, transporters, metabolic enzymes, and their affiliated proteins) or from the degradation of aberrant proteins via P3 (proteases and their affiliated proteins). The former is defined as AA supply flux (v1, J1) and the latter as the degradation flux (v3, J3). AAs are translated into polypeptides via R (ribosome-affiliated proteins), defined as translation flux (v0, J0). Unfolded polypeptides (UP) mature as native proteins via P2 (chaperones and other proteins promoting protein maturation) or mistakenly mature as aberrant proteins (AP) because of the lack of P2. The former is defined as normal maturation flux (v2, J2) and the latter as aberrant maturation flux (v4, J4). APs can also form protein aggregates spontaneously, defined as the aggregation flux (v5, J5). Here, vi denotes the total flux rate and Ji (, where Mc denotes total cell mass and i ∈ {0, 1, 2, 3, 4, 5}) denotes flux rate per unit cell mass. Each flux can be represented as a function of the concentration or proteome fraction of related proteins.