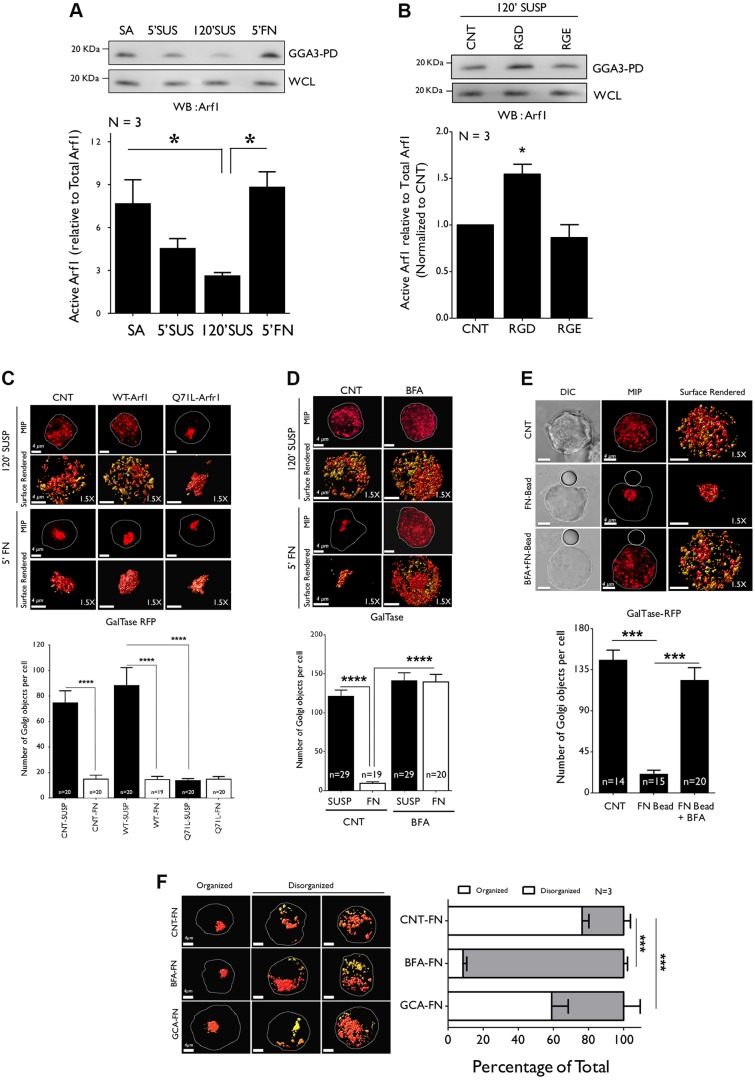
Fig. 5.
Adhesion-dependent Arf1 activation regulates Golgi organization. (A) Western blot detection of activated Arf1 (WB: Arf1) immunostained in GST-GGA3 (GGA3 PD) and total Arf1 in whole-cell lysate (WCL) from stable adherent (SA), detached (5′ SUS), suspended for 120 min (120′ SUS) and re-adherent (5′ FN) WT-MEFs. (B) Western blot of activated Arf1 in GST-GGA3 pulldown (GGA3-PD) and of total Arf1 in WCL of WT-MEFs suspended for 120 min (120′ SUSP) and mock treated with the volume equivalent of Milli-Q water (CNT), or treated with RGD peptide (RGD) or RGE peptide (RGE) in Milli-Q water for 15 min. The graphs in A and B represent the densitometric band intensity ratio of activated Arf1 (GGA3-PD) to total Arf1 as the mean±s.e. from 3 independent experiments. (C) WT-MEFs transfected with GalTase alone (CNT), GFP-tagged WT-Arf1 or constitutively active Q71L Arf1 were suspended for 120 min (120′ SUSP) and re-plated on FN for 5 min (5′ FN). (D) GalTase expressing WT-MEFs suspended for 90 min were treated with methanol as a control (CNT) or (10 µg/ml) BFA for 30 min and re-plated on FN for 5 min (5′ FN) without or with BFA. (E) GalTase-expressing WT-MEFs were suspended for 90 min and incubated for 30 min without beads as control (CNT) or with beads coated with FN and incubated with methanol (FN-Bead) or BFA (BFA+FN-Bead). In all of the above, representative MIP and surface-rendered de-convoluted images (1.5× magnified) of cells are shown, together with DIC images in the FN-bead study. Graphs represent discontinuous Golgi objects per cell as the mean±s.e. of 15–29 cells (as indicated in graph) from 3 independent experiments. (F) WT-MEFs expressing GalTase suspended for 60 min were treated with 10 µg/ml DMSO (mock), 10 µM BFA or 10 µM Golgicide-A (GCA) for 30 min and re-plated on FN alone (CNT-FN) or FN with BFA (BFA-FN) or FN with GCA (GCA-FN). The distribution of cells with organized and disorganized Golgi phenotypes in these populations was determined (in percent) and representative surface-rendered cross-section images are shown. The graph represents mean±s.e. from 3 independent experiments. Scale bars in images is 4 µm. Statistical analysis was done using Mann–Whitney U test and Chi-Square test for distribution profile (*P<0.05, **P<0.001, ***P<0.0001, ns=non-significant).