Figure 4. The GRA is under the control of the P. aeruginosa LASI/RHLI and IQS QS systems.
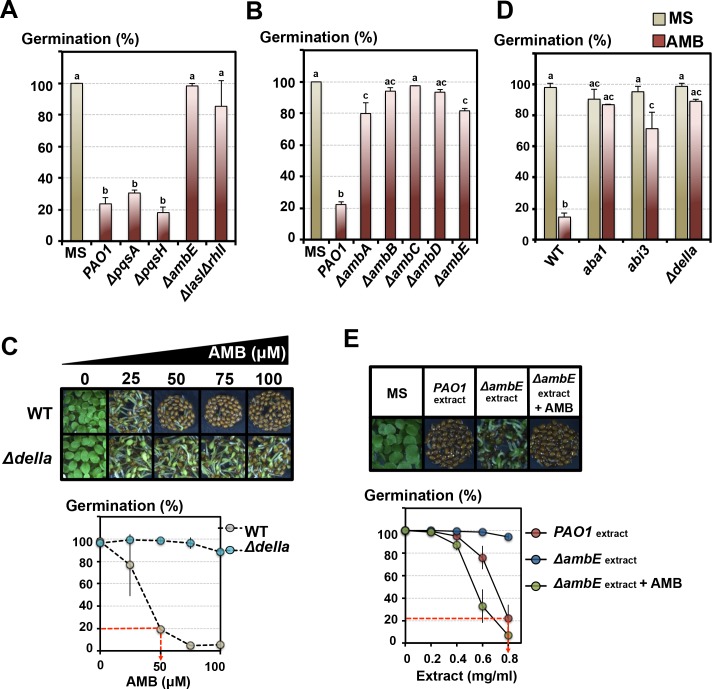
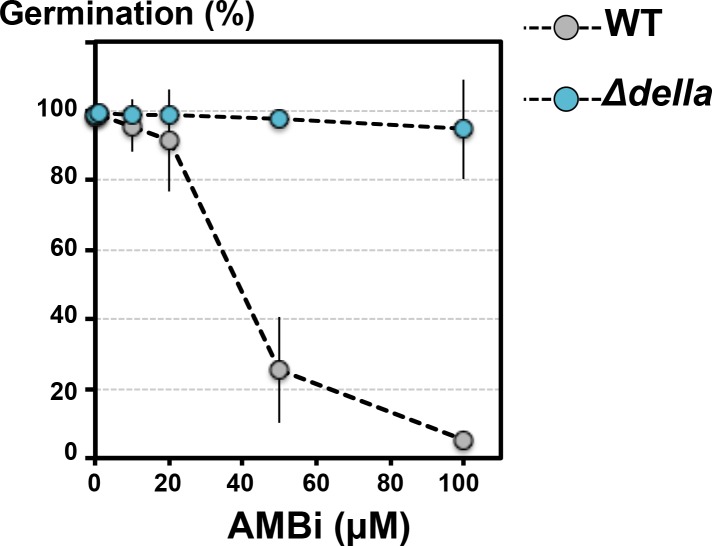
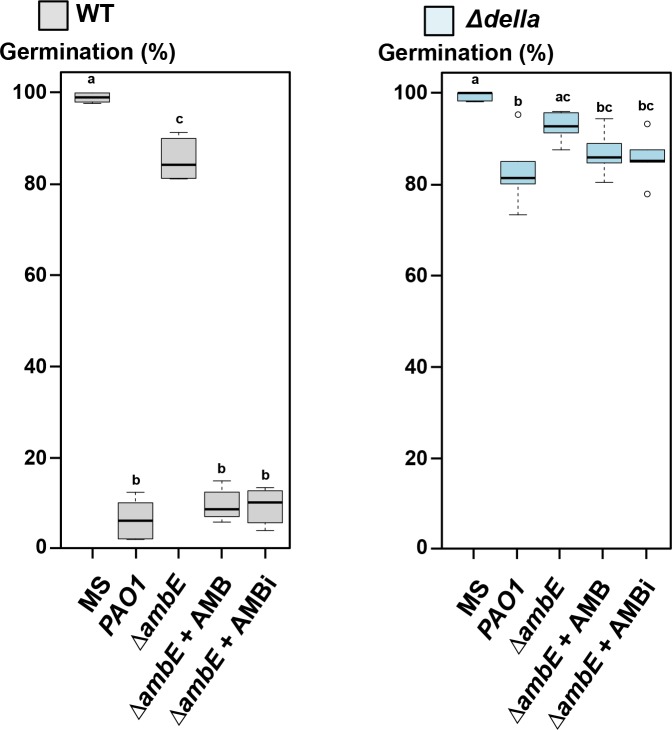
L-2-amino-4-methoxy-trans-3-butenoic acid (AMB) is the main GRA released by P. aeruginosa. (A) Histograms show seed germination percentage 4 days upon sowing seeds in absence (MS) or presence of WT P. aeruginosa (PAO1) or mutant P. aeruginosa strains as indicated. PAO1 is the reference WT P. aeruginosa strain from which all the QS mutant strains were derived. Seeds and P. aeruginosa cells were separated by 2 cm. Data represent mean ± standard deviation (four replicates, n = 150–200). Statistical treatment and lower case letters as in Figure 1B. (B) Same experiment as in A. using P. aeruginosa ΔambA-E mutants, as indicated, each affected in individual AMB operon genes. Data represent mean ± standard deviation (three replicates, n = 100–150). Statistical treatment and lower case letters as in Figure 1B. (C) AMB represses Arabidopsis seed germination in a DELLA-dependent manner. Pictures show representative Arabidopsis plants 4 days after sowing WT and Δdella seeds in presence of different AMB concentrations as indicated. The graph shows quantification of the germination percentage of WT and Δdella seeds exposed to AMB. The red dashed line indicates the concentration of synthetic AMB (50 µM) having the same GRA as that present in germination plates containing 0.8 mg/ml of PAO1 extracts (see Figure 4E). Data represent mean ± standard deviation (three replicates, n = 150–200). (D) AMB represses Arabidopsis seed germination in an ABA-dependent manner. Histograms show germination percentage of WT, aba1, abi3 and Δdella 3 days after sowing seeds in absence (MS) or presence of 50 µM of synthetic AMB. Data represent mean ± standard deviation (two replicates, n = 100–150). Statistical treatment and lower case letters as in Figure 1B. (E) Synthetic AMB introduces a GRA in ΔambE extracts equivalent to that of PAO1 extracts. ΔambE extracts, which lack AMB, were supplemented with synthetic AMB so as to obtain the same amount of AMB naturally present in PAO1 extracts. Pictures show representative Arabidopsis plants 4 days after sowing WT seeds in absence (MS) or presence of WT P. aeruginosa extracts (PAO1extract) or ΔambE P. aeruginosa extracts (ΔambEextract) or AMB-supplemented ΔambE extracts (ΔambEextract + AMB). 0.8 mg/ml of WT P. aeruginosa extract was used. The graph shows quantification of the germination percentage of WT seeds exposed to the different extract concentrations as indicated. Note that the supplemented ΔambE extract (ΔambEextract + AMB) has a GRA equivalent, if not higher, than that of PAO1 extract. The red dashed line indicates the concentration of PAO1 extract (0.8 mg/ml) having the same GRA as that present in germination plates containing 50 µM synthetic AMB (see Figure 4C). Data represent mean ± standard deviation (six replicates, n = 250–300).
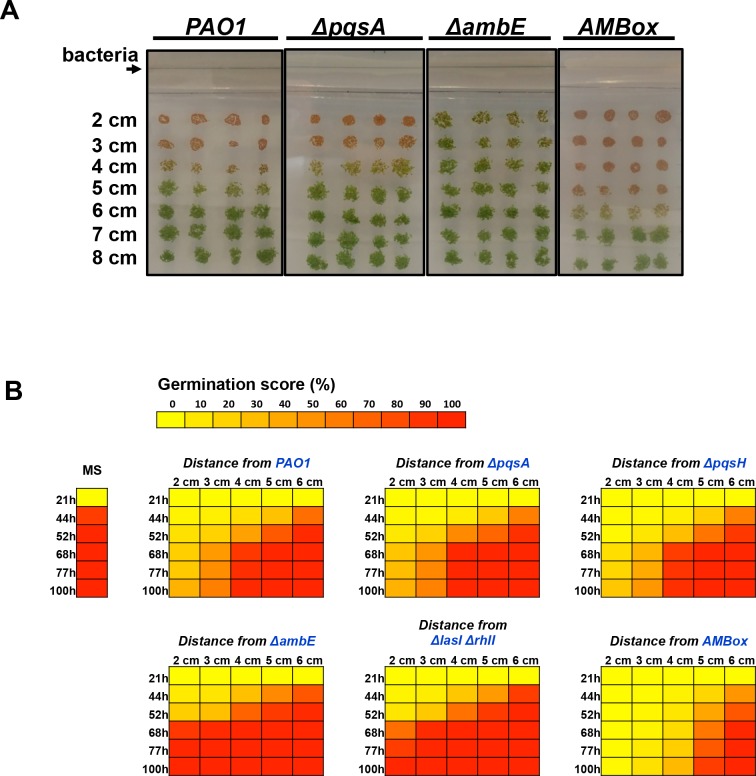
Figure 4—figure supplement 1. The GRA is under the control of the P. aeruginosa LASI/RHLI and IQS QS systems.
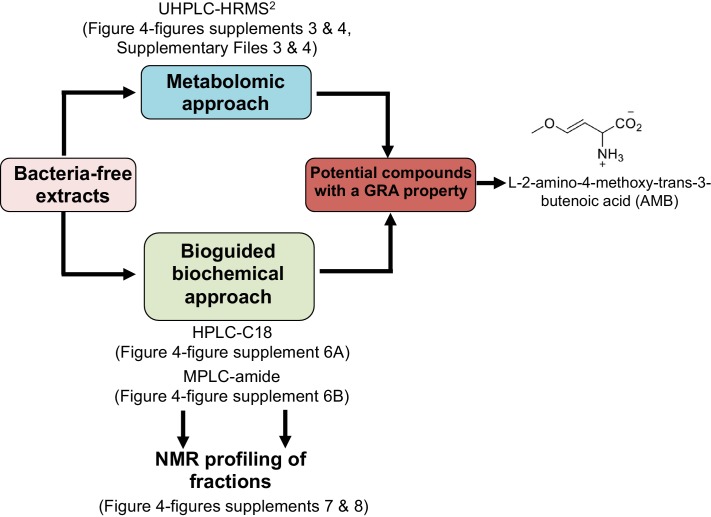
Figure 4—figure supplement 2. Approaches used in this study to identify compounds released by P. aeruginosa with a germination repressive activity (GRA).
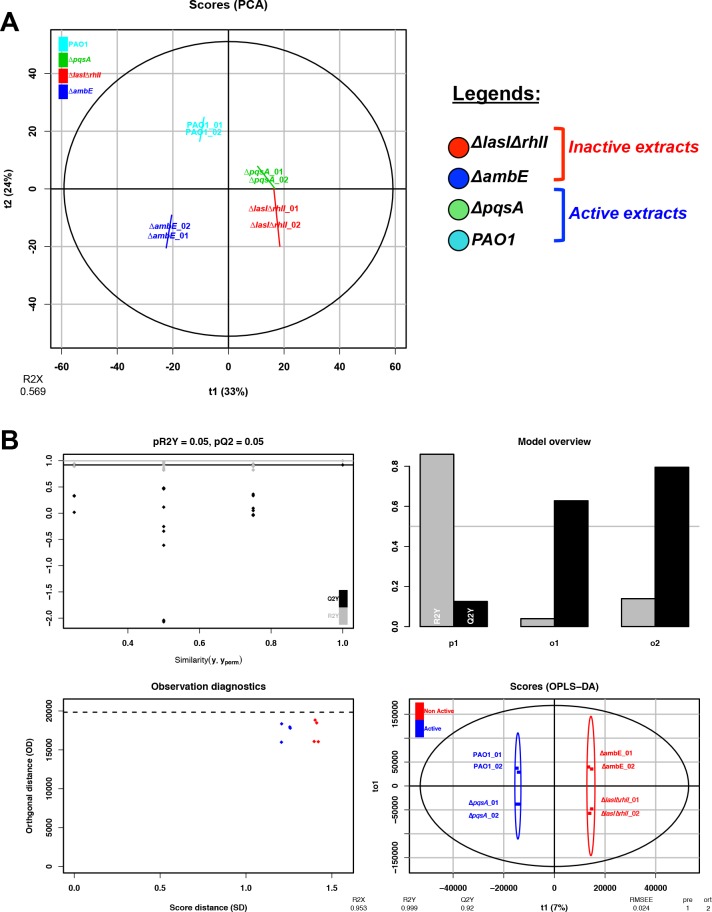
Figure 4—figure supplement 3. Unsupervised and OPLS-DA results from metabolomic analysis 1.
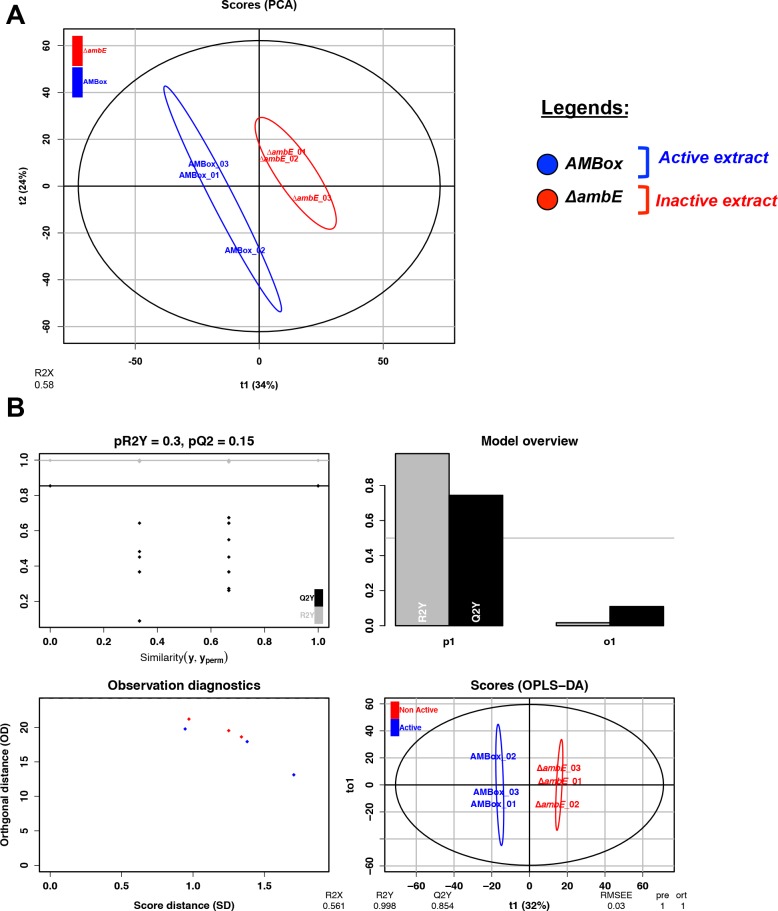
Figure 4—figure supplement 4. Unsupervised and OPLS-DA results from metabolomic analysis 2.
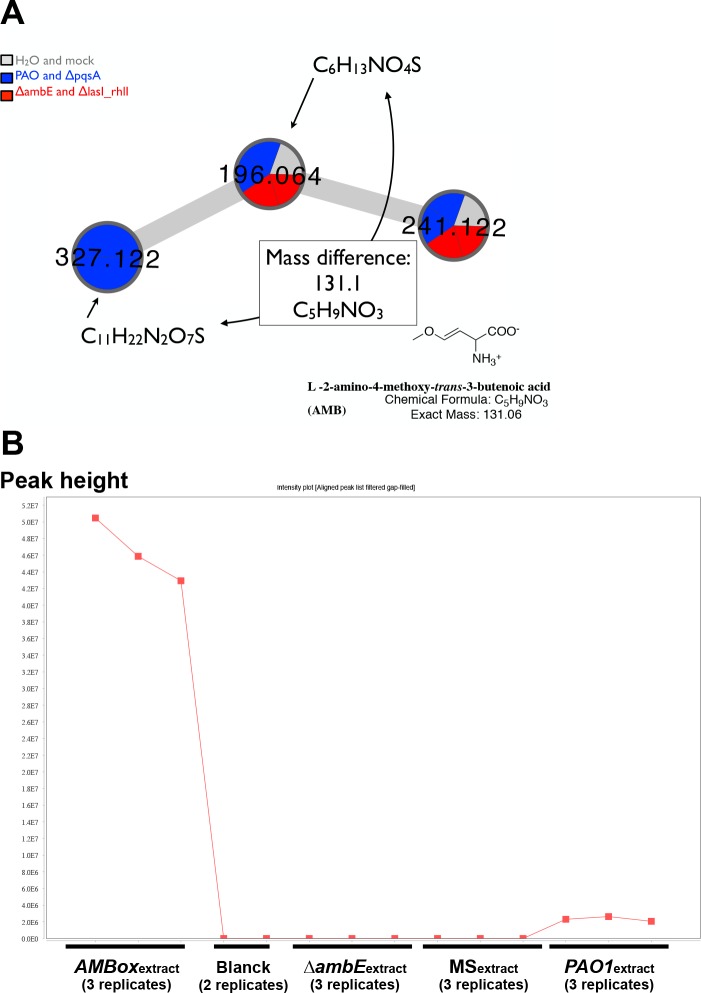
Figure 4—figure supplement 5. m/z 327.12 is the Mass Spectrometry (MS) signature of AMB.
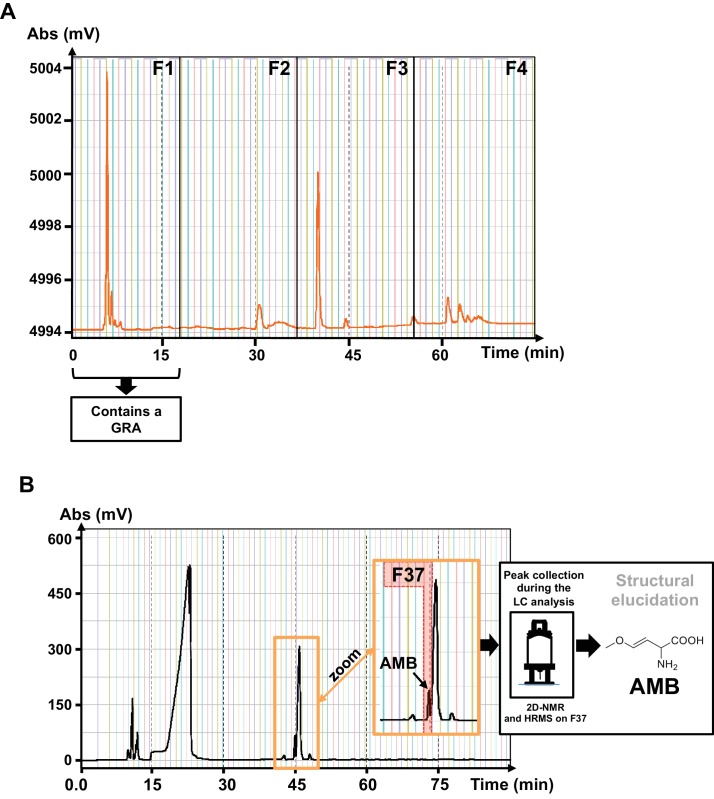
Figure 4—figure supplement 6. Isolation of the GRA released by P.aeruginosa.
Figure 4—figure supplement 7. Fraction F37 contains AMB and a GRA.
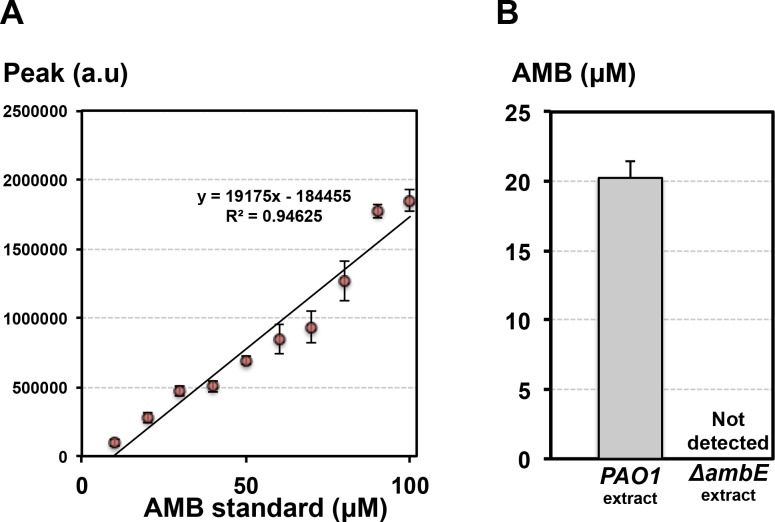
Figure 4—figure supplement 8. Quantification of AMB in Pseudomonas extracts.
Figure 4—figure supplement 9. ∆ambE extracts supplemented with AMBi or synthetic AMB contain the same GRA.
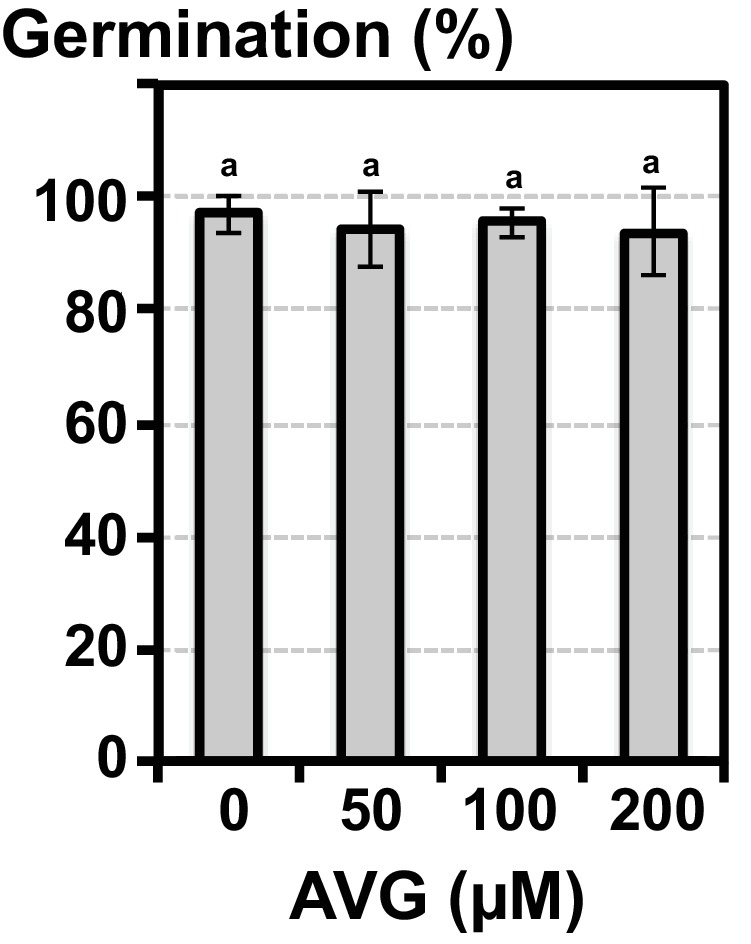
Figure 4—figure supplement 10. The oxyvinylglycine AVG does not inhibit Arabidopsis seed germination.