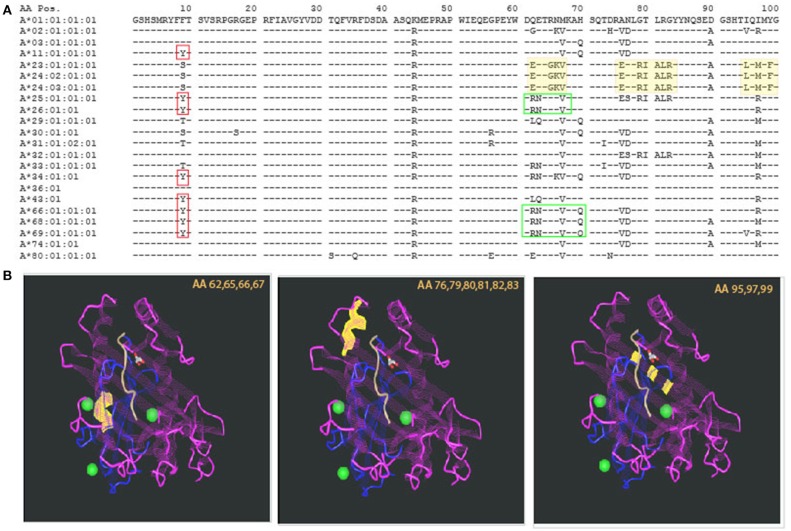
Figure 1.
Polymorphism and homology in the HLA system. (A) The sequence of the first 100 amino acids of HLA-A alleles is presented using the one letter naming convention. Allele identity is listed to the left. Consensus sequence is represented by the sequence of HLA-A*01:01:01:01. For additional sequences, homology to the consensus sequence is illustrated by a dash sign (“−”). The vast majority of the other alleles' sequences are homologous to the consensus sequence. Polymorphism is represented by a single letter designation of the different amino acid. Even among polymorphic positions, there is identity with some of the other alleles. This is demonstrated by the red boxes—all of those alleles have Y in position 9 but other alleles are identical to the consensus sequence (F) and yet others have (S). Other examples are illustrated by the green boxes. (B) Polymorphism can be distributed in different areas of the HLA molecule. Three examples are illustrated by the yellow highlights in the sequence, and the corresponding sites of the molecule are shown in the 3 insets, listing the polymorphic amino acids that are highlighted in yellow. Those are located, from left to right: at the lower edge of the alpha helix, around center molecule; the alpha helix at the edge of the peptide binding groove; and at the bottom of the peptide binding groove—beta pleated sheets. The projected effects on T cell receptor and the bound peptide are likely to change based on the location of the polymorphism.