Figure 2.
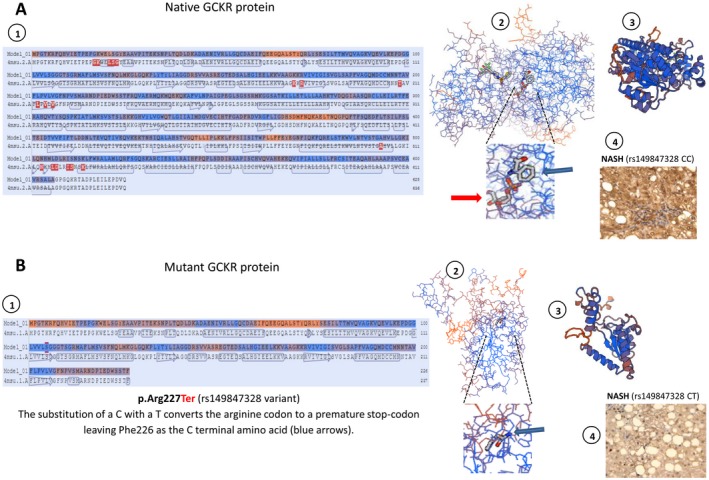
Protein structure modeling of the p.Arg227Ter mutation in the GCKR gene. Prediction was built using the SWISS‐MODEL re(https://swissmodel.expasy.org/). (A) Representation of native GCKR protein and (B) mutant protein (stop gained) showing structural changes as the result of a premature stop codon in residue 227. A1 and B1 show amino acid sequence alignment of native and mutant protein, respectively. A2 and B2 show the backbone protein structure; insets highlight the effect of the p.Arg227Ter mutation that is responsible for a loss in the binding of a sugar (D‐sorbitol‐6‐phosphate 9, red arrows); its binding sites involve several amino acids downstream of Phe226 that are then lost in the mutated protein (B2). A3 and B3 show a three‐dimensional ribbon diagram of wild‐type and mutant protein conformational structures, respectively; p.Arg227Ter truncates the protein length by about two thirds. A4 and B4 show representative protein expression patterns of GCKR in the liver of a patient with NASH who carries two copies of the wild‐type (rs149847328 CC) and one copy of the mutant (rs149847328 CT) allele, respectively. A4 shows positive staining in more than 50% of cells; B4 shows positive staining in less than 20% of cells. Liver protein expression of GCKR was evaluated using immunohistochemistry; immunostaining for GCKR was performed on a subsample of six liver specimens previously included in paraffin (four samples of patients with NASH and similar phenotypic and clinical features of the affected patient and two liver specimens of the affected patient, including baseline and follow‐up liver biopsies). Immunoreactivity was examined using light microscopy of liver sections; original magnification ×400.
