Figure 1.
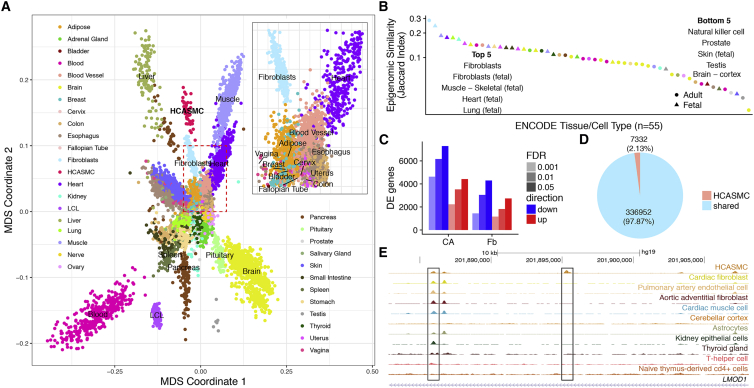
The Relationship between HCASMCs and GTEx and ENCODE Cell and Tissue Types
(A) The multidimensional scaling plot of gene expression shows that HCASMCs form a distinct cluster, which neighbors fibroblast, skeletal muscle, heart, blood vessel, and various types of smooth muscle tissues such as esophagus and vagina (inset).
(B) Jaccard similarity index between HCASMCs and ENCODE cell and tissue types reveals that fibroblast, skeletal muscle, heart, and lung are most closely related to HCASMCs.
(C) Thousands of genes are differentially expressed between HCASMCs and its close neighbors, fibroblast, as well as the tissue of origin, coronary artery.
(D) A total of 344,284 open chromatin peaks are found in HCASMCs, of which 7,332 (2.1%) are HCASMC specific.
(E) An example of a HCASMC-specific peak located within the intron of LMOD1, which is an HCASMC-specific gene.