Figure 2.
Tissue- and Cell Type-Specific Contribution to CAD Risk
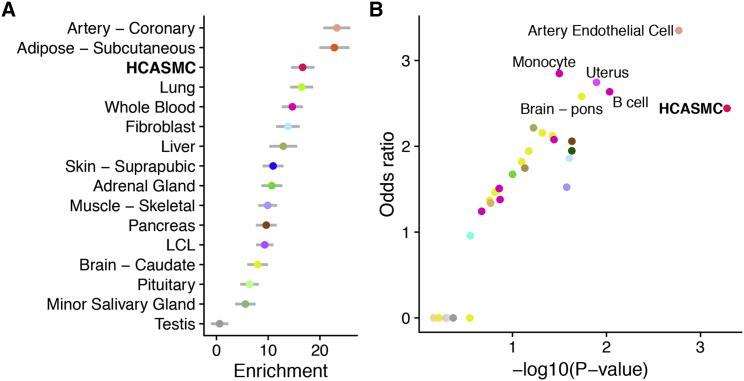
(A) Tissue-specific enrichment of CAD heritability. We used stratified LD score regression to estimate the CAD risk explained by SNPs close to tissue-specific genes, defined as the 2,000 genes with highest expression z-scores (see Material and Methods). Genes whose expression is specific to coronary artery, adipose, and HCASMCs harbor SNPs with large effects on CAD. Error bars indicate standard error of the enrichments.
(B) Overlap between CAD risk variants and tissue- and cell type-specific open chromatin regions. We used a modified version of GREGOR (see Material and Methods) to estimate the probability and odds ratio (compared with matched background SNPs) of overlap between CAD risk variants and open chromatin regions in HCASMCs and across ENCODE tissues. HCASMCs, arterial endothelial cells, monocytes, B cell, uterus (composed primarily of smooth muscle), and pons (possibly through regulation of blood pressure) showed the highest degrees of overlap.