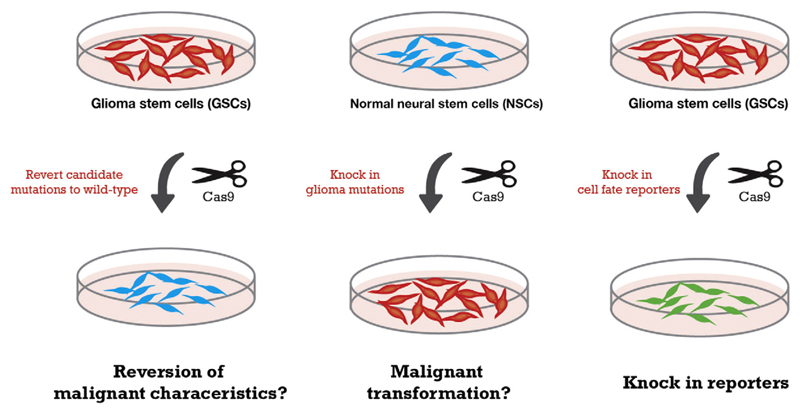
Fig. 2.
A genome editing ‘toolkit’ for functional genetic studies and novel engineered cellular models. Reversion of candidate drivers to wild-type (left) and/or introduction of key drivers into normal NSCs (middle) using CRISPR/Cas9 provides both a means for proving mutation causality, and matched cell lines as perfect isogenic controls for drug screening. Creation of a variety of useful engineered alleles such as live cell reporters and safe-harbours (right) removes need for fluorescent staining. Co-culture of fluorescently tagged cell lines carrying potentially druggable driver mutations with isogenic non-tagged controls provides an ideal internally controlled cell assay.