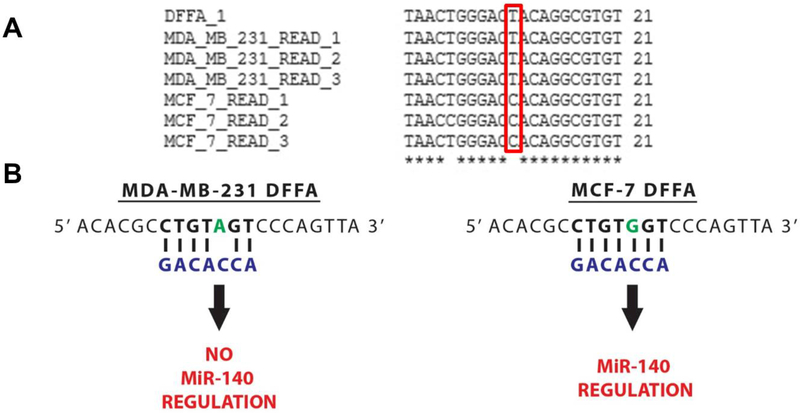
Figure 5.
MiR-140 can regulate DFFA in MCF-7, but not MDA-MB-231. (A) Alignment of 21 nt segments of six RNA-Seq reads (three from each cell line) to a portion of the apoptosis inducing gene DFFA. Our edit identification algorithm identified an A-to-G edit site at basepair 10,460,668 on Chromosome 1, and corresponding reads mapping to that location were extracted and trimmed to 21 bp (edit site plus/minus 10 bp flanking regions). Edit location is outlined in red. The alignment was generated via ClustalW [40]. (B) Illustration showing complimentary base pairing between the miR-140 seed (blue) and the DFFA gene in both cell lines. The edit site is indicated in green.