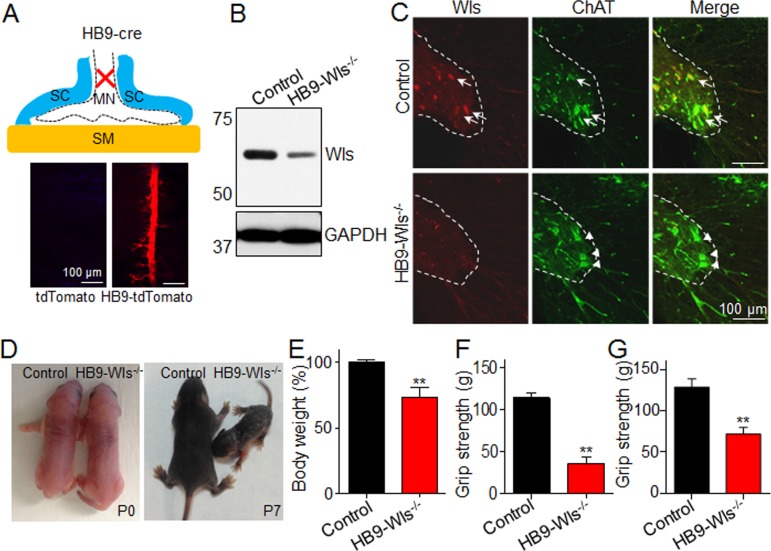
Figure 2. Ablation of Wls expression in motoneuron causes muscle weakness.
(A) Up: NMJ structure. Cre targets motoneuron (HB9::cre). Down: Cre expression was verified by crossing HB9::cre mice with Rosa-tdTomato reporter mice (HB9-tdTomato). HB9::cre drives tdTomato expression in the middle line of diaphragm where motoneuron innervates in the muscle (HB9-tdTamato, (P7). MN, motoneuron; SC, Schwann cells; SM, skeletal muscle. (B) Immunoblot of Wls in sciatic nerve from 2-month-old control and HB9-Wls-/- mutant. Notice that the reduced Wls protein expression in the mutant. (C) Immunostaining of Wls in spinal cord from 2-month-old control and HB9-Wls-/- mutant. Notice that Wls staining signal is lost in the ChAT positive cells in the spinal ventral horn in the HB9-Wls-/-. Anti-Wls (Red); Anti-ChAT (Green). Arrows indicate colocalization of Wls and ChAT protein. (D) Two genotypes at P0 and P7. Notice that apparent normal size at P0, and reduced body size at P7. (E) Body weight of two genotypes at P7. Control, n = 20; mutant, n = 11; Unpaired t-test, **p<0.01. (F) Grip strength of two genotypes at P60. Control, n = 5; mutant, n = 5. Unpaired t-test, **p<0.01. (G) Grip strength of two genotypes with same body weight at P60. Control, n = 5; mutant, n = 5. Unpaired t-test, **p<0.01.