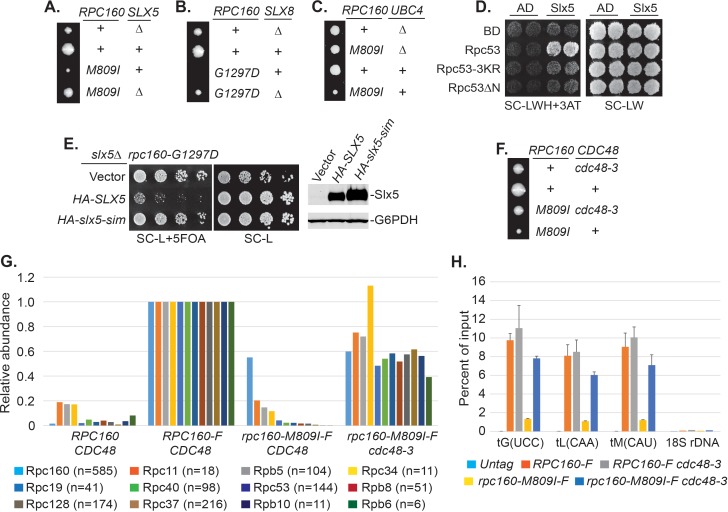
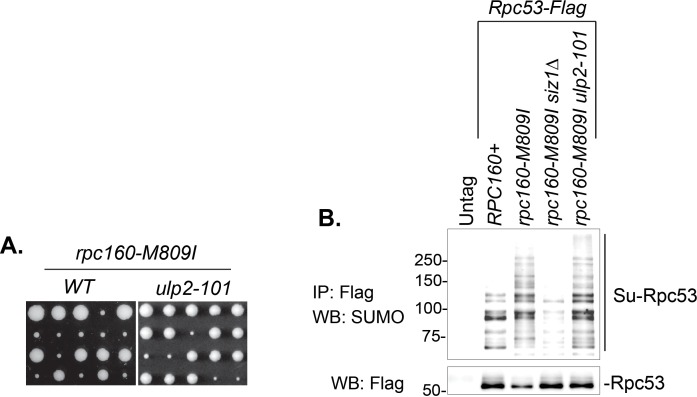
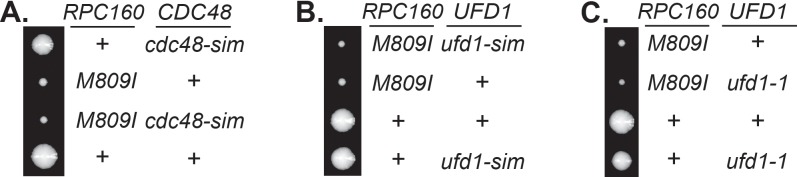
Figure 5. Pol III is repressed by ubiquitylation and p97/Cdc48.
(A–C) The indicated rpc160 mutant strains were crossed with slx5Δ, slx8Δ, or ubc4Δ strain, respectively, followed by tetrad analysis. The cross between slx8Δ and rpc160-G1297D was shown, because slx8Δ caused obvious growth defect by itself, so the rescue effect was more obvious on rpc160-G1297D, which is a sicker mutant than rpc160-M809I. (D) Yeast two-hybrid interactions between Slx5 and Rpc53. SLX5 and RPC53 were cloned into a 2μ LEU2 Gal4 activation-domain (AD) vector and a 2μ TRP1 DNA-binding domain (BD) vector, respectively, and co-transformed into yeast strain PJ69-4A. Transformants were selected on synthetic media lacking leucine and tryptophan (SC-LW), then patched and replica plated to selective media lacking histidine to test for interactions. The histidine-lacking media was supplement with 3-aminotriazole (SC-LWH + 3AT) for a more stringent phenotype. (E) LEU2 plasmids carrying HA-tagged wild type or SIM-defective SLX5 (HA-slx5-sim) were transformed into an rpc160-G1297D slx5Δ strain containing a URA3 RPC160 plasmid. Transformants were selected on SC-L then spotted onto an SC-L + 5 FOA plate to lose wild-type RPC160. HA-SLX5 complemented slx5Δ so the cells became sicker compared to the empty vector control transformants, while HA-slx5-sim did not complement, indicating that the SIMs are essential for the function of SLX5 in this assay. The lost Slx5 function by the SIM mutations was not caused by insufficient proteins, since there were comparable levels of Slx5 proteins, as determined by an anti-HA immunoblot on total cell lysates (right, top panel). G6PDH served as a loading control (right, bottom panel). (F) Tetrad analysis between rpc160-M809I and cdc48-3. (G) Determination of Rpc160 association with other Pol III subunits. Pol III complexes containing Flag-tagged wild type or mutant Rpc160 in wild-type CDC48 or cdc48-3 cells were isolated using anti-Flag agarose beads, followed by TMS labeling and mass-spec analysis to quantify the relative amounts of Rpc160-interacting proteins. Signals for 12 of the 17 Pol III subunits, including Rpc160, were detected, and normalized to the signals from RPC160-Flag cells. n = Number of times when a unique peptide for the indicated protein is measured. An untagged RPC160 strain was used as a negative control. (H) ChIP analysis of Rpc160. Flag-tagged RPC160 or rpc160-M809I was expressed from a plasmid in wild-type CDC48 or cdc48-3 cells, as indicated. The Flag-tagged proteins were purified using anti-Flag agarose beads. An untagged strain was used as negative control. Three tRNA gene loci, as well as 18S rDNA (negative control), were examined. Chromatin association was determined by real-time PCR of the indicated genomic loci, using the percent of input method. Data are mean ± standard deviation calculated from six data points (two biological replicates and three technical replicates).