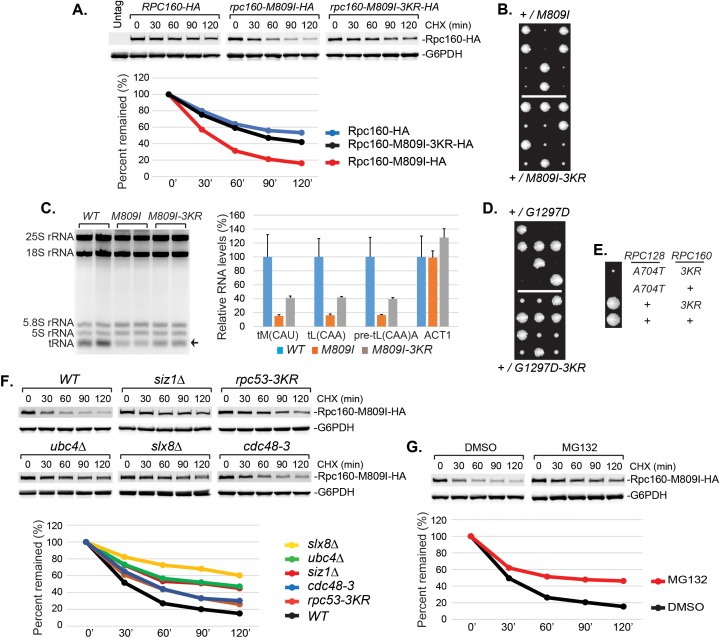
Figure 6. Pol III repression by ubiquitylation is partially mediated through Rpc160.
(A) CEN URA3 plasmids expressing HA-tagged wild type or mutant Rpc160, as indicated, were transformed into a wild-type strain, and their stabilities were assayed during a cycloheximide (CHX) chase time course. Rpc160 was detected by an anti-HA antibody, and G6PDH was used as a loading control. Quantification of the bands was shown below the immunoblot. (B) Tetrad analysis of the diploid strains, RPC160+/rpc160-M809I (top) and RPC160+/rpc160-M809I-3KR (bottom). Tetrads from these two diploids were dissected and plated on the same YPD plate at the same time, in order to compare the growth of rpc160-M809I and rpc160-M809I-3KR cells. The growth of three dissected tetrads were shown. The large colonies are wild-type RPC160 cells, and the small colonies are rpc160-M809I (top) or rpc160-M809I-3KR (bottom) cells. The rpc160-M809I-3KR cells grew slightly faster than the rpc160-M809I cells. (C) Left: 2 μg of RNA extracted from the indicated strains was run on a 2.8% agarose gel containing ethidium bromide, then visualized with UV. Two colonies were picked for each strain. Right: RNA from left was reverse transcribed into cDNA, followed by real-time PCR analysis, as described in Figure 2E. GAPDH transcripts were used as loading control. (D) Similar tetrad analysis as in (B) of the diploid strains, RPC160+/rpc160-G1297D (top) and RPC160+/rpc160-G1297D-3KR (bottom). Large colonies are wild-type RPC160 cells, while the missing colonies (top) are rpc160-G1297D cells, and the small colonies (bottom) are rpc160-G1297D-3KR cells. (E) An rpc128-A704T strain was crossed with an rpc160-3KR strain, followed by tetrad analysis. (F) A CEN URA3 rpc160-M809I-HA plasmid was transformed into the indicated strains, and the stabilities of the Rpc160-M809I-HA proteins were determined by CHX chase time course, as described in (A). (G) A CEN URA3 rpc160-M809I-HA plasmid was transformed into a wild-type strain, and protein stabilities were determined by CHX chase experiment in the presence of DMSO or MG132.