Figure 2: XLF-XRCC4 interaction is required for SR-complex formation.
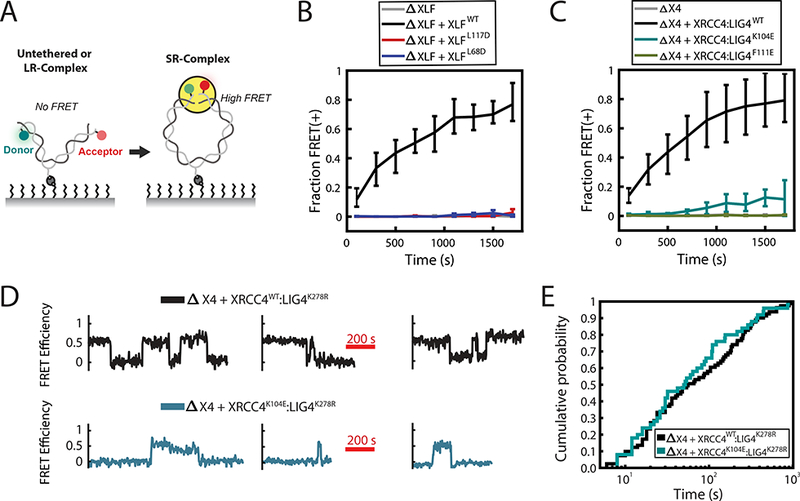
(A) Schematic of intramolecular single-molecule FRET reporter for monitoring short-range synaptic complex (SR-complex) formation38. DNA is tethered internally via a biotin-streptavidin interaction and labeled 7 bp from each end with Cy3 (Donor) and Cy5 (Acceptor) fluorophores. No FRET is detected when DNA ends are untethered or in the long-range synaptic complex (LR-complex). Close alignment of DNA ends within the SR-complex is indicated by energy transfer between Cy3 and Cy5. The data shown in B and C were collected using a 0.5 s exposure time, alternating between two frames of Cy3 excitation and one frame of Cy5 excitation. The data shown in D and E were collected using a 1 s exposure with a 1 s delay between exposures, alternating between four frames of Cy3 excitation and one frame of Cy5 excitation.
(B-C) Kinetics of SR-complex formation in extract depleted of XLF or XRCC4 and supplemented with purified recombinant protein, as in Fig. 1C-D. The mean fraction of FRET-positive (SR-complex or ligated) substrates is plotted as a function of time after extract addition. Error bars represent the minimum and maximum values obtained from multiple experimental replicates. See Supplementary Table 2 for sample sizes.
(D) Sample smFRET trajectories showing SR-complex formation and dissociation in XRCC4-immunodepleted extract supplemented with either XRCC4WT:LIG4K278R or XRCC4K104E:LIG4K278R. Catalytically inactive LIG4K278R was used here and in panel E to prevent ligation of DNA ends.
(E) Cumulative distribution functions of SR-complex lifetimes. The difference between the two conditions was not statistically significant (p = 0.45, log-rank test). See Supplementary Table 3 for sample sizes.
