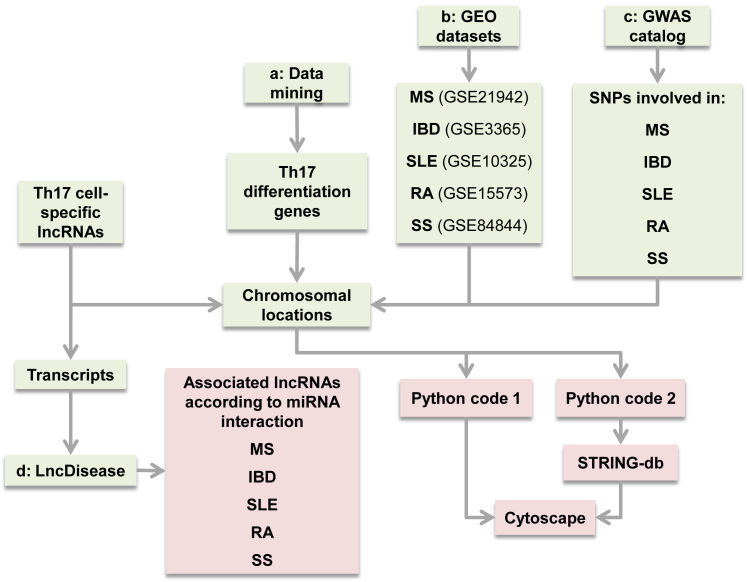
Figure 10.
In Silico Workflow of the Study
Lineage-specific lncRNAs were obtained from RNA-seq data. In step a, SNPs associated with each AID were retrieved from the GWAS catalog for analysis of their proximities to lncRNAs through Python programming language and were then visualized in Cytoscape. In step b, differentially expressed mRNAs in AID patients were obtained from corresponding GEO datasets. After chromosomal localization, their distances from selected lncRNAs were checked via Python programming language and, after inputting to STRING-db, visualized in Cytoscape. In step c, Th17 cell differentiation genes were selected through data mining. Their neighboring to the selected lncRNAs was determined and, after inputting to STRING-db, visualized in Cytoscape. In step d, lncRNAs’ transcripts were deduced and inputted to the LncDisease software, which checked lncRNA-miRNA interaction and outputted the diseases related to the predicted miRNAs.