Fig. 8.
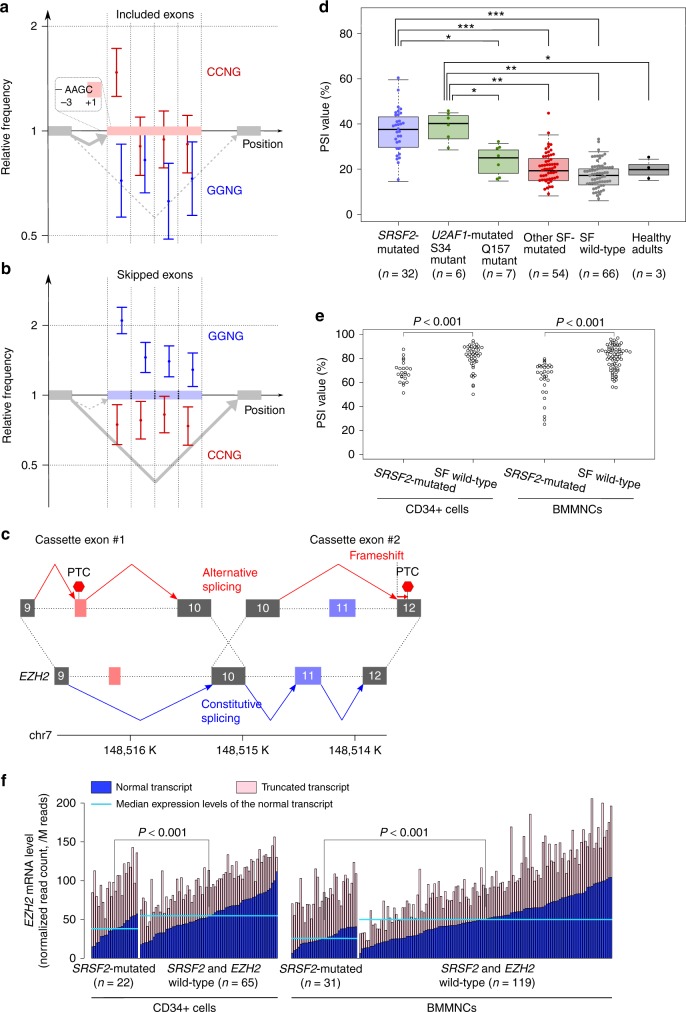
Mutant SRSF2-associated Cassette Exons in EZH2. a, b Forest plots show relative frequencies of CCNG and GGNG motifs within the differentially spliced exons in the SRSF2-mutated samples. Two panels show the exons that are more frequently included (n = 380) (a) and skipped (n = 645) (b) in the SRSF2-mutated samples, respectively. Comparisons were made between the differentially spliced exons and constitutive ones (n = 72,002). Red and blue dots indicate relative frequencies of CCNG and GGNG, respectively. Bars indicate 95% confidence intervals. Relative frequencies are shown for exons divided into four segments with equal lengths. c Two cassette exons in EZH2 associated with SRSF2 mutation. The left is alternative exon usage and the right is exon skipping. Gray boxes indicate constitutive exons. Red and blue boxes denote exons with increased and decreased usage in the SRSF2-mutated samples, respectively. d PSI values of the cryptic exon between exons 9 and 10 of EZH2. BMMNC samples with different SF mutation status are plotted separately. The middle line in each box corresponds to the median; the lower and upper boundaries of the box indicate first and third quartiles, respectively. The whiskers represent the 1.5-fold interquartile range or the maximum/minimum data point within the range. *** indicates Bonferroni adjusted P value < 0.001; **, adjusted P value < 0.01; *, adjusted P value < 0.05. e PSI values of exon 11 of EZH2. SRSF2-mutated samples and those without SF mutations are plotted separately for BMMNCs and CD34+ cells. The number of samples in each group is shown in Fig. 1c. f mRNA levels of EZH2 isoforms compared between the SRSF2-mutated samples and those without SRSF2 and EZH2 alterations. One sample with both SRSF2 and EZH2 mutations were removed from this analysis. Normal transcripts are those from an EZH2 isoform without SRSF2-associated alterations in exon usage. Truncated transcripts are those from EZH2 isoforms with at least one SRSF2-associated altered exon usage. Levels of normal transcripts were compared using the Mann–Whitney U test. W-values were 340 in CD34+ cells and 630 in BMMNCs