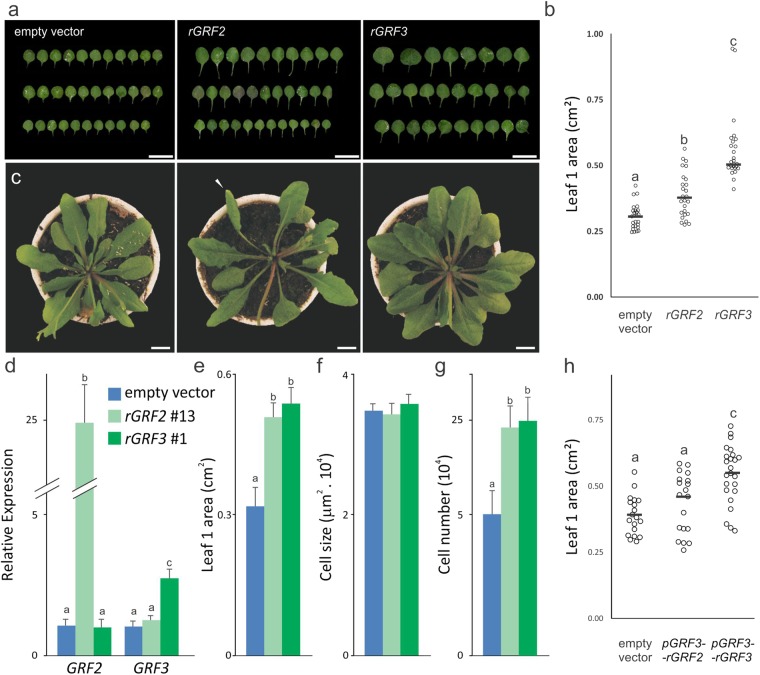
Figure 3.
Superior capacity of rGRF3 compared to rGRF2 in increasing leaf size. (a,b) Fully expanded leaf 1 size distribution in a population of Arabidopsis primary transgenic plants transformed with the empty vector, rGRF2 or rGRF3. In the scatterplots in panel b, each circle indicates the size of a single leaf from an independent T1 transgenic plant, while the horizontal solid bar represents the sample median. Different letters indicate significant differences, as determined by ANOVA followed by Tukey’s multiple comparison test (P < 0.05). Bars = 1 cm. (c) 30-days old plants transformed with the empty vector, rGRF2 or rGRF3. Note the leaf shape changes induced by rGRF2 only (arrowhead), including long and twisted petioles with downward curled leaves. Bars = 1 cm. (d) Expression levels of At-GRF2 and At-GRF3 in homozygous T3 transgenic plants transformed with the empty vector or rGRF2 (rGRF2 #13) or rGRF3 (rGRF3 #1). The GRF levels were estimated by RT-qPCR and normalized to control plants (empty vector). The data shown are mean ± SEM of three biological replicates. Different letters indicate significant differences as determined by ANOVA followed by Tukey’s multiple comparison test (P < 0.05). (e–g) Leaf 1 size (e), palisade cell size (f) and estimated palisade cell number (g) in selected rGRF2 #13 and rGRF3 #1plants. Different letters indicate significant differences as determined by ANOVA followed by Tukey’s multiple comparison test (P < 0.05). (h) Fully expanded leaf 1 size distribution in a population of primary transgenic Arabidopsis plants transformed with rGRF2 or rGRF3 under the GRF3 promoter. In the scatterplots each circle indicates the size of a single leaf from an independent T1 transgenic plant, while the horizontal solid bar represents the sample median. Different letters indicate significant differences, as determined by ANOVA followed by Tukey’s multiple comparison test (P < 0.05). Bars = 1 cm.