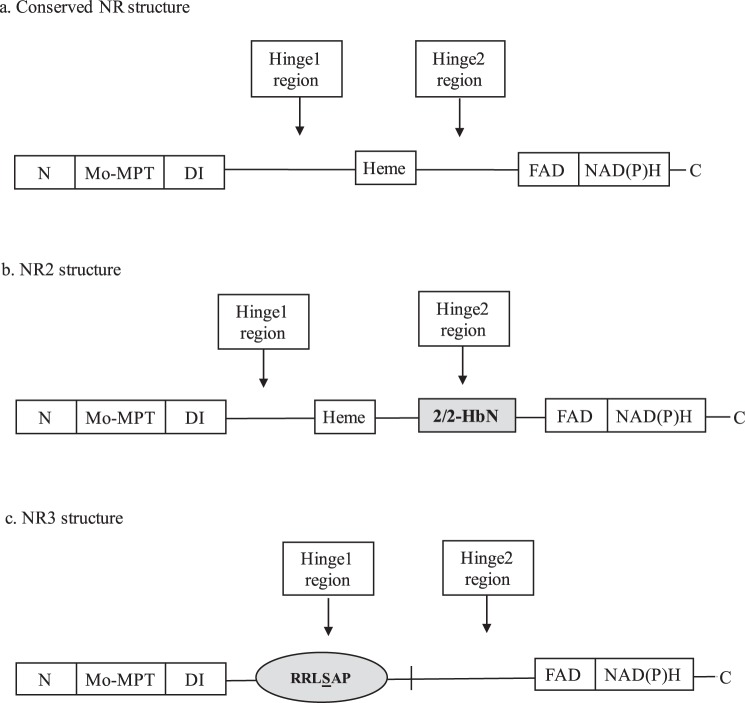
Figure 1.
Alternative domain structures for nitrate reductase enzymes. (a) The conserved structure of nitrate reductase in higher plants, algae, and fungi, depicting five domains (MO-MPT, DI, Heme-Fe, FAD, and NADH domains) and 3 regions (N-terminal, hinge 1 and hinge 2 regions)3. (b) The structure of NR2 with a 2/2HbN domain (indicated by grey box) inserted in its Hinge 2 region of Heterosigma akashiwo and Chattonella subsalsa2. (c) The structure of NR3 lacking the Heme-Fe domain and with a 14-3-3 protein binding motif (indicated by grey circle) at a phospho-Ser residue (underlined in RRLSAP) in the Hinge 1 region of Chattonella subsalsa. N: N-terminal region; Mo-MPT: molybdenum-molybdopterin cofactor domain; DI: dimer interface; Heme: cytochrome b5-binding domain containing Heme-Fe; FAD: flavin adenine dinucleotide domain; NAD(P)H: NAD(P)H domain; C: C-terminal of NR enzyme.