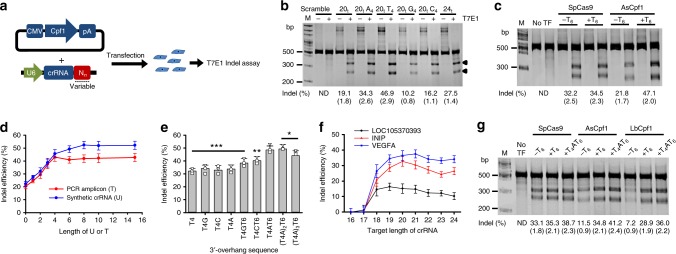
Fig. 2.
Optimized configuration of crRNA for highly efficient genome editing in vivo. a Scheme of the in vivo assay to determine the most efficient configuration of the crRNA. Cpf1-encoding plasmids were co-transfected with crRNA-encoding PCR amplicons into HEK-293T cells, and the Cpf1 activity was assessed by T7E1 indel assays. b Improved indel efficiency of AsCpf1 in vivo by the U-rich 3′-overhang following a 20-nt target-complementary sequence in transcribed crRNAs. This gel image is a representative result of three repeated experiments. Indel values are mean±standard deviation. c Improved indel efficiency by the 3′-end U-rich guide RNA as a unique feature of Cpf1. 3′-Proximal addition of uridinylates did not change the indel efficiency of SpCas9 in vivo. This gel image is representative of three repeated experiments. Indel values are the mean±standard deviation. d Improved indel efficiency of AsCpf1 in vivo by increased uridinylate lengths. The AsCpf1 activity was improved by the increased lengths of 3′-end uridinylates up to 8–10 mers for the chemically synthesized crRNA and up to six bases for the crRNA-encoding PCR amplicons. e Optimized 3′-end configuration of crRNA for highly efficient genome editing. Addition of the U4AU4 3′-overhang in crRNA maximized the indel efficiency of AsCpf1. *p > 0.05, **p < 0.05, ***p < 0.01 (n = 3), two-tailed Student’s t test. f The optimal target length for the use of a U-rich crRNA. A target length of 20 (±1) nt was optimal for the U-rich crRNA. g Validation of the optimal crRNA configuration for highly efficient genome editing using CRISPR-Cpf1. The optimal configuration of the U4AU4 3′-overhang in addition to a 20-nt target-matched sequence was identically applied for LbCpf1 as well as AsCpf1, but not for SpCas9. This gel image is representative of three repeated experiments. Indel values are the mean±standard deviation