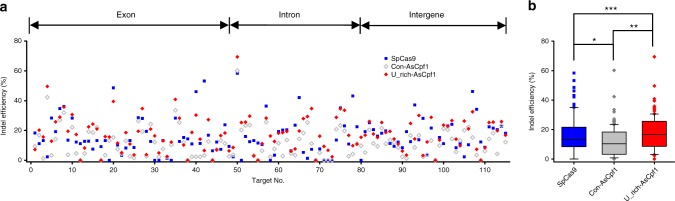
Fig. 3.
Large-scale comparison of the genome-editing efficiencies of CRISPR-AsCpf1 and CRISPR-SpCas9. a A dot plot of the indel efficiencies of AsCpf1 and SpCas9 in HEK-293T cells. The indel efficiencies of AsCpf1 and SpCas9 were compared on common targets with a sequence of 5′-TTTV(N)20NGG-3′, where V is A, C, or G. For the AsCpf1 activity, the conventional crRNA (for con-AsCpf1) configuration was compared with our optimized U-rich crRNA (for Opt-AsCpf1). b A box-and-whisker plot for the indel efficiencies of AsCpf1 and SpCas9. The graph consists of median values, lower and upper quartile, and outer standard deviations. *p = 0.003, **p = 0.00003, ***p = 0.29, two-tailed Student’s t test