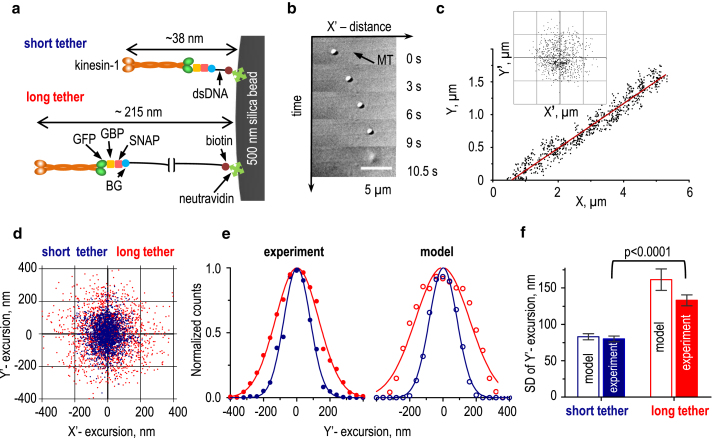
Figure 2.
Verification of the TCM assay using kinesin-1 and dsDNA links. (a) Our strategy for conjugation of kinesin-1 to the surface of a glass bead via dsDNA links of different lengths. The SNAP-GBP protein adaptor enables connection of this GFP-labeled motor to the BG-labeled dsDNA. (b) Selected DIC images are shown of a bead transported by kinesin-1 linked via the long dsDNA link. The bead detached 10.5 s after the start of motion. (c) Experimental coordinates were collected for a bead conjugated to kinesin-1 via the long dsDNA link. The inset shows a cloud of excursions for this bead, calculated as the deviations of the bead’s position from the line representing the MT (MT-perpendicular Y’-excursion) and from the assumed motor attachment site (MT-parallel X’-excursion); see Supporting Materials and Methods. The grid size is 0.2 μm. (d) Experimental cloud plots are shown for microbeads conjugated to kinesin-1 with different dsDNA links; representative data sets are based on the total 12,000 coordinates collected for 29 beads with the short tethers and 15,000 coordinates for 56 beads with the long tethers (for clarity, only 2000 randomly selected coordinates are shown for each tether). MT-parallel X’-excursions for these plots were calculated using the sliding averaging windows of 100–300 time points, corresponding to 1–3 s. (e) Normalized histograms show distributions of experimentally measured versus modeled bead Y’-excursions for the short (blue) and long (red) tethers. Each data set was fitted to a Gaussian function and normalized to its mean value. Each distribution is based on 2000 coordinates. (f) SDs of bead Y’-excursions are shown. Model predictions are based on data in Fig. 1e. Experimental data were obtained from 29 beads with short tethers and 56 beads with long tethers. Error bars are SDs generated by bootstrap analysis; p-values were calculated by unpaired t-test. To see this figure in color, go online.