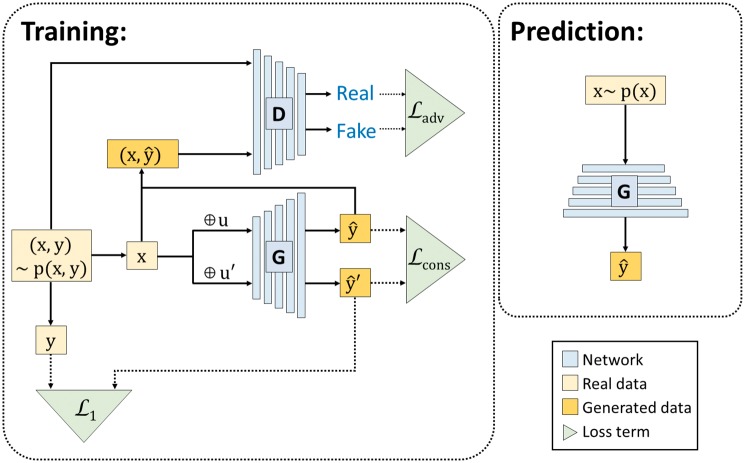
Fig. 1.
Illustration of GGAN architecture and its loss functions. We use to denote a gene expression profile, where corresponds to the landmark genes and represents the target genes. Our goal is to learn a generator function G which takes x as the input and output as the prediction of the target gene expression. To construct an appropriate prediction function G, we consider three loss terms in our model: and . measures the consistency of the prediction from G when the input x is perturbed by random noise u and . measures the difference between the prediction vector and the ground truth y. For the term , we construct a discriminator D which takes both and as the input. The discriminator D tries to distinguish the real’ sample from the ‘fake’ sample while the G tries to predict the realistic vector to fool the discriminator. measures the adversarial loss in the game between the generator G and discriminator D