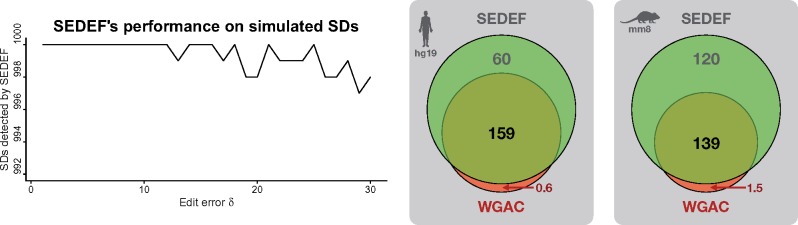
Fig. 3.
(Left) Performance of SEDEF’s algorithm on simulated SDs. x-axis is the total simulated SD error rate δ, whereas y axis is the number of correctly detected SDs (total 1000 for each δ). Since SEDEF successfully detects more than 995 simulated SDs for any δ, the plot area is cropped. (Right) Venn diagram depicts the SD coverage of the human and mouse genome (in Mbp) as calculated by SEDEF and WGAC. Intersected region stands for the bases covered by both SEDEF and WGAC