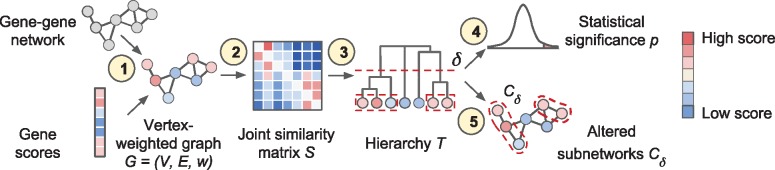
Fig. 2.
Overview of the Hierarchical HotNet method. (1) A biological interaction network and gene scores are paired to form a vertex-weighted graph . (2) A joint similarity matrix S is derived from both network topology and vertex weights using a random walk-based approach. (3) A dendrogram T of vertex sets is constructed from S using asymmetric hierarchical clustering. (4) The statistical significance p of T is evaluated (test statistic and null model for G not shown). (5) Representative altered subnetworks are provided from the dendrogram T